A Fun Introductory Command Line Lesson: Next Generation Sequencing Quality Analysis with Emoji!
Editor: Srebrenka Robic
Published online:
Abstract
Radical innovations in DNA sequencing technology over the past decade have created an increased need for computational bioinformatics analyses in the 21st century STEM workforce. Recent evidence however demonstrates that there are significant barriers to teaching these skills at the undergraduate level including lack of faculty training, lack of student interest in bioinformatics, lack of vetted teaching materials, and overly full curricula. To this end, the James Madison University, Center for Genome & Metagenome Studies (JMU CGEMS) and other PUI collaborators are devoted to developing and disseminating engaging bioinformatics teaching materials specifically designed for streamlined integration into general undergraduate biology curriculum. Here, we have developed and integrated a fun introductory level lesson to command line next generation sequencing (NGS) analysis into a large enrollment core biology course. This one-off activity takes a crucial but mundane aspect of NGS quality control (QC) analysis and incorporates the use of Emoji data outputs using the software FASTQE to pique student interest. This amusing command line analysis is subsequently paired with a more rigorous research-grade software package called FASTP in which students complete sequence QC and filtering using a few simple commands. Collectively, this short lesson provides novice-level faculty and students an engaging entry point to learning basic genomics command line programming skills as a gateway to more complex and elaborated applications of computational bioinformatics analyses.
Primary image: Undergraduate students learn the basics of command line NGS quality analysis using the FASTQE and FASTP programs.
Citation
St. Jacques RM, Maza WM, Robertson SD, Lonsdale A, Murray CS, Williams JJ, Enke RA. 2021. A fun introductory command line lesson: Next generation sequencing quality analysis with Emoji! CourseSource. https://doi.org/10.24918/cs.2021.17
Society Learning Goals
Bioinformatics
- Computation in the life sciences
- What is the role of computation in hypothesis-driven discovery processes within the life sciences?
- What computational concepts are important in bioinformatics?
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
- Computational Skills
- What higher-level computational skills can be used in bioinformatics research?
Lesson Learning Goals
Students will:
- Appreciate how Next Generation Sequencing (NGS) data supports biological research.
- Learn the importance of the FASTQ file format and quality control in NGS Data Analysis.
- Understand how basic command line scripts and bioinformatics tools are used to assess and enhance the quality of NGS FASTQ files.
Lesson Learning Objectives
Students will be able to...
- Write basic command line scripts and use simple command-line bioinformatics tools.
- Explain how next generation sequencing data is used to generate and address hypotheses in biological research.
- Discuss the importance of assessing the quality of sequencing data.
- Identify the components of a FASTQ file.
- Assess sequencing read quality using command-line to generate FASTQE and FASTP reports.
- Trim and filter low quality reads in a FASTQ file.
- Compare and contrast FASTQE and FASTP tools.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
The rise of Big Data acquisition and analytics in the 21st century continues to permeate into a growing number of professional domains. Since the advent of commercial Next Generation Sequencing (NGS) technology about 12 years ago, genomics data represents one of the most proliferative Big Data domains with unprecedented growth projected by the year 2025 (1). Exponential growth of the genomics Big Data domain has already begun to shift the needs of the modern STEM workforce with an increased demand for combined training in the life sciences as well as computational bioinformatics (2). A recent nation-wide survey study of biology faculty in the US conducted by the Network for Integrating Bioinformatics into Life Sciences Education (NIBLSE) group identified nine consensus bioinformatics core competencies recommended for ubiquitous integration into undergraduate life sciences education (3). However, significant barriers to integrating these competencies have been well documented including lack of faculty training, lack of student interest in bioinformatics, lack of vetted teaching materials, and overly full curricula (4). Furthermore, a disproportionately lower number of faculty teaching at minority-serving institutions (MSIs) report integration of bioinformatics into their teaching compared to faculty at non-MSIs (4).
Several notable examples of courses have been developed to address bioinformatics analysis in undergraduate curriculum. These courses, however, typically require extensive faculty expertise in bioinformatics to implement (5-6), require large portions of instructional time devoted to bioinformatics analysis (7-10), or focus mainly on web-based point and click introductory bioinformatics tools rather than more advanced programming-based analysis (11,12). To help overcome these barriers, a variety of open access resources have also been developed such as the GOBLET training portal and the QUBES platform (13,14). These platforms offer educators a variety of well-vetted introductory bioinformatics materials available for free download and facile implementation into a variety of teaching contexts. Even with these available resources, there is a shortage of introductory materials to introduce computer scripting applicable to life sciences, particularly in large enrollment introductory courses.
Here we report on the development and integration of a unique in-class introduction to genomics command line coding lesson that reinforces several of the NIBLSE core competencies while also surmounting reported barriers to integration. This short, one-off lesson is designed to fit into a single class period aimed at both novice students and educators with little to no coding experience. Additionally, to pique student interest in the methodology and subject matter, the lesson takes a crucial but typically mundane aspect of NGS quality control (QC) analysis and incorporates amusing Emoji data outputs in place of typical QC output metrics using the command line software package FASTQE (https://fastqe.com). FASTQE analysis is followed by more rigorous research-grade analysis in which students complete sequence QC and filtering with the command line software package FASTP (15; https://github.com/OpenGene/fastp). Collectively, this short lesson provides novice-level students and faculty an engaging entry point to learn basic genomics command line coding skills as gateway to stacking more complex principles and applications of computational bioinformatics analyses. Our implementation of this lesson into a large enrollment 200 level Genetics Lab course provided the opportunity to introduce command line scripting to a variety of students across academic years from multiple academic majors.
Intended Audience
This lesson can be effective for a wide variety of audiences, including life science undergraduates and faculty with a baseline knowledge of DNA Biology and next generation sequencing (NGS) techniques but who are novices to command line scripting. Here, we focus on implementation of this lesson into a large enrollment core curriculum laboratory course at a 4-year public school (James Madison University). Bio 240 Genetics is a 200 level course required for all JMU Biology, Biotechnology, and Health Sciences majors. During the fall 2019 semester, this course enrolled 278 students all of whom had taken at least two introductory biology courses as prerequisites. The course is usually taken during the 2nd year of the Biology/Biotechnology core curriculum sequence, but also serves juniors and seniors in other majors as well. Ideally, this short lesson can also be incorporated into other life science courses to supplement student learning of modern NGS technology as well as to introduce concepts in computer programming at scale early in student's undergraduate careers. Additionally, this lesson can be effectively implemented in a traditional in person lab as well as a synchronous or asynchronous online formatted course.
Required Learning Time
This lesson is designed for a 45-60 minute class period. Incorporation of optional extended learning questions will increase the length of the lesson by approximately 15-30 minutes, however this portion of the activity can be assigned as either an in-class or take-home assignment or removed altogether based on instructor preference. A short 20-30 minute pre class assignment is also necessary to ensure that students have the required computing capabilities prior to class if they are using their own personal computers for the lesson. Refer to Table 1 for more details on lesson timing. An alternative implementation using a cloud-based Jupyter Notebook that does not require additional software installation is also available (Supporting File S6. FASTQE – Jupyter Notebook alternative implementation instructions).
Prerequisite Student Knowledge
This lesson focuses on teaching basic command line coding to analyze genomics NGS data. A basic understanding of DNA biology is recommended for students (i.e., a full year of introductory biology). No prior experience with command line scripting is required as this lesson is aimed at novices; however, a basic understanding of Illumina or other similar NGS techniques and data output is suggested for most effective learning.
Prerequisite Teacher Knowledge
This lesson assumes that the instructor has some baseline knowledge of genomics and NGS technology. Prior experience with command line scripting is not required, though it is recommended that the instructor be comfortable enough with the materials to help students troubleshoot any technical issues with their command line scripts. By completing the lesson prior to implementation, instructors will be able to test their level of comfort.
SCIENTIFIC TEACHING THEMES
Active Learning
During the in-class lesson, students can work either individually or in groups to follow a step-by-step tutorial and actively explore how to use Linux commands and view FASTQ data with Emoji. Students participate in hands-on exercises and work on the corresponding problems alternatively and at their own pace. The problems are intentionally inserted into the tutorial. Peer and/or instructor guidance and discussions are provided when needed. Extended learning questions are available for students pursuing advanced learning.
Assessment
Students are assessed primarily via successful completion of required tasks and the scores received by answering questions inserted into the tutorial. An answer key is provided in the instructor version of the activity (Supporting File S7. FASTQE – Instructor version of lesson).
Inclusive Teaching
To help students with diverse learning methods achieve a complete understanding, lesson materials are presented in multiple ways: traditional lecture with visual aids combined with a group hands-on learning activity. The hands-on activity is supplemented with formative assessment questions in which students reflect on the activity to formulate answers based on their in-class analysis. Furthermore, nation-wide survey data indicates that barriers to integrating bioinformatics into undergraduate life sciences curriculum are disproportionately greater to faculty teaching at minority-serving institutions (MSIs) compared to faculty at non-MSIs (4). This disparity increases the value of engaging STEM undergraduates in computational bioinformatics, where there is evidence of a diversity gap. Additionally, this lesson can be effectively implemented in a traditional in person lab as well as a synchronous or asynchronous online formatted course.
LESSON PLAN
Student Pre-Class Preparation
The degree of required student pre-class preparation depends on the computers that will be used for the lesson. Ideally, classroom computers preloaded with required software are provided to groups or pairs of students. In this instance no pre-class preparation is required. If students use their own computers however, they will need to complete a short pre-class assignment ensuring that an Anaconda software bundle required for the command line lesson is downloaded and installed on their machines (Supporting File S1. FASTQE – Pre-class assignment) This activity requires a high-speed internet connection and should take 20-30 minutes to complete (Table 1). It should be noted that students using their own computers typically raises the probability of encountering user-specific problems with the lesson that may be more difficult to troubleshoot (Table 2). An alternative implementation using a cloud-based Jupyter Notebook that does not require additional software installation is also available (Supporting File S6. FASTQE – Jupyter Notebook alternative implementation instructions).
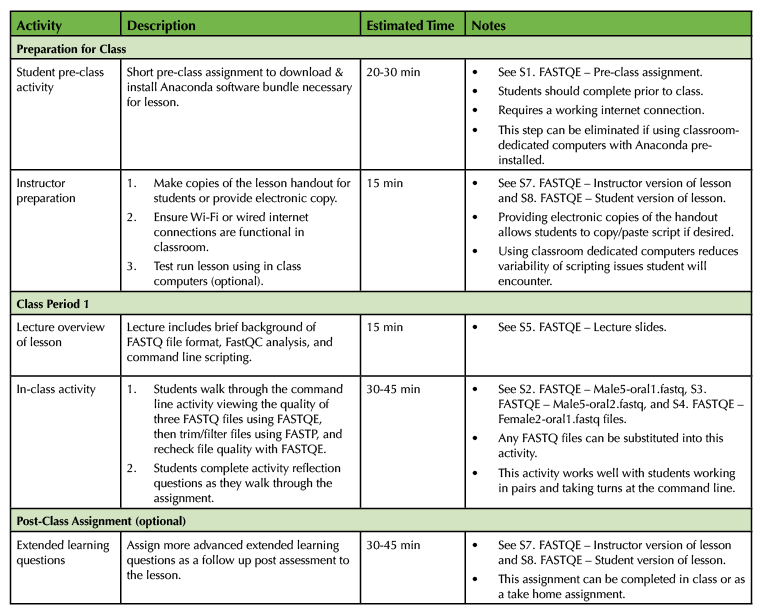
Table 1. Introduction to Command Line Scripting Lesson Timeline.
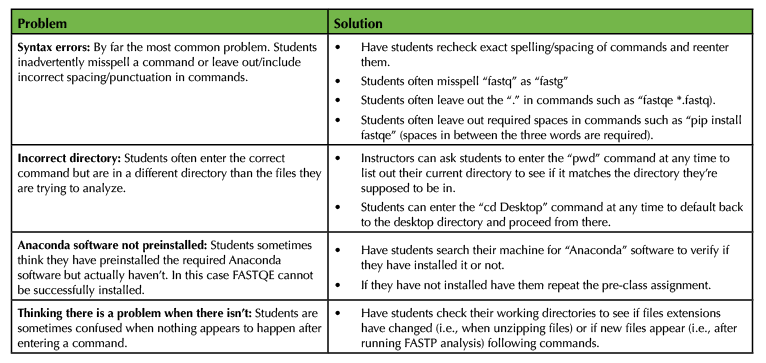
Table 2. Common mistakes and pitfalls of FASTQE-FASTP activity.
Instructor Pre-Class Preparation
Instructors should also prepare by completing the pre-class assignment to ensure that all required software are installed on the computer they will use for the lesson (Supporting File S1. FASTQE – Pre-class assignment). Ideally, this will be a Wi-Fi enabled laptop allowing the instructor to move around the classroom to demonstrate and troubleshoot lesson specifics during the instruction period. Alternatively, an internet-ready stationary computer connected to an overhead projector will suffice. To best prepare for our command line lesson, instructors should run through the entire activity in advance of the class. To do this, instructors will need to download the three provided FASTQ files (Supporting File S2. FASTQE – Male5-oral1.fastq file, Supporting File S3. FASTQE – Male5-oral2.fastq file, and Supporting File S4. FASTQE – Female2-oral1.fastq file) or alternative FASTQ files for analysis. An instructor version of the lesson provides solutions to the activity question and problems (Supporting File S7. FASTQE – Instructor version of lesson). If classroom computers will be provided for the lesson, instructors may choose to download required software and FASTQ files on individual machines to eliminate the need for the student pre-class assignment. If student computers will be used, instructors will need to make zipped FASTQ files available for download via institutional learning management systems (i.e., Canvas, Blackboard, etc.) or Cloud storage (i.e., GoogleDrive, Microsoft OneDrive, etc.). Alternatively, a Jupyter Notebook web-based implantation of this lesson can be used with classroom and/or student computers in which FASTQ files are bundled and do not need to be provided by instructors (Supporting File S6. FASTQE – Jupyter Notebook alternative implementation instructions). Instructors should also print copies of the student version of the assignment to hand out to each individual student participating in the lesson (Supporting File S8. FASTQE – Student version of lesson). Table 2 outlines the most common problems encountered with this lesson and tips for troubleshooting.
In-Class Command Line Lesson
Prior to the hands-on activity, a short slide-assisted lecture is presented to orient students to background information on FASTQ file format, NGS quality analysis, and FASTQE software (for lecture slides see Supporting File S5. FASTQE – Lecture slides). The lecture explains how NGS sequencers such as Illumina and Ion Torrent rely on Phred probability scores for each individual base call in a sequencing read to determine the read's accuracy (Figure 1A). Since NGS sequencers typically read millions-billions of sequences per sample simultaneously, the popular software package FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) is commonly used to visualize the quality of all reads in a sample by plotting the average Phred score at each base position to create a Per Base Quality plot (Figure 1B). The novel software package FASTQE (https://fastqe.com) performs a similar analysis to FastQC; however, rather than plotting the average Phred score at each base position, an emoji symbol correlated with a quality score is plotted (Figure 1C). Following the introductory lecture, students work in pairs or small groups at their own pace walking through the hands-on activity (Supporting File S8. FASTQE – Student version of lesson). FASTQE software equates each Phred score between 0-41 to an individual emoji symbol (Figure 1D).
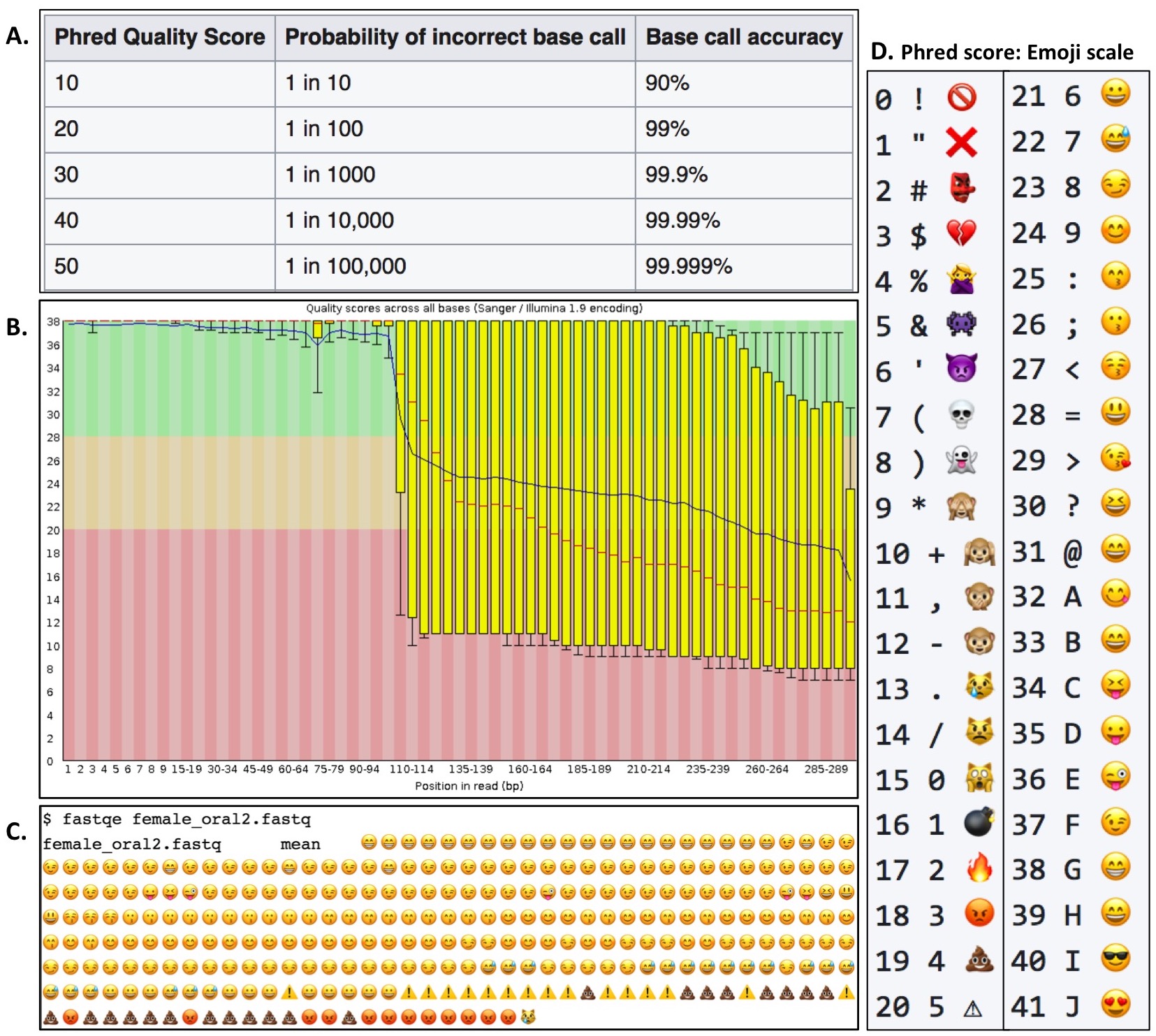
Figure 1. Overview of the command line program FASTQE. A Phred quality score designates the probability of accuracy for each individual base call produced by DNA sequencers (A). Example of a FastQC Per Base Quality plot for the female_oral2.fastq file (B). Example FASTQE plot of the same FASTQ file (C). FASTQE equates an emoji output associated with the averaged Phred score at each base position of the sequencing read (D).
During FASTQE analysis, students output amusing and somewhat informative Per Base Quality plots visualizing a FASTQ file's overall quality (Figure 2A). Due to their relevance in popular culture, students intuitively understand the implication of various emoji symbols and their relationship to base call quality at a given position. FASTQE analysis certainly lacks research grade rigor but is very effective at engaging students in the practice of scripting and understanding NGS quality analysis. Introducing FASTQE is therefore an effective primer for having students then move onto more rigorous command line analysis in the second part of the activity. After visualizing their Per Base Quality plots in FASTQE, students then use a second command line program called FASTP (https://github.com/OpenGene/fastp) to trim and filter FASTQ files. FASTP is a research grade bioinformatics tool that removes low quality sequence reads and base calls from a FASTQ file. With a single command, FASTP filters FASTQ files outputting pre and post filtered Per Base Quality plots for each file analyzed as well as a filtered FASTQ file for subsequent analysis (Figure 2B). Students then rerun FASTP-filtered FASTQ files again in FASTQE to generate filtered versions of the emoji Per Base Quality plot to compare to pre filtered versions (Figure 2C). Collectively, this short in class activity takes novice students from introductory command line scripts through research-grade command line bioinformatics analysis in a single class period.
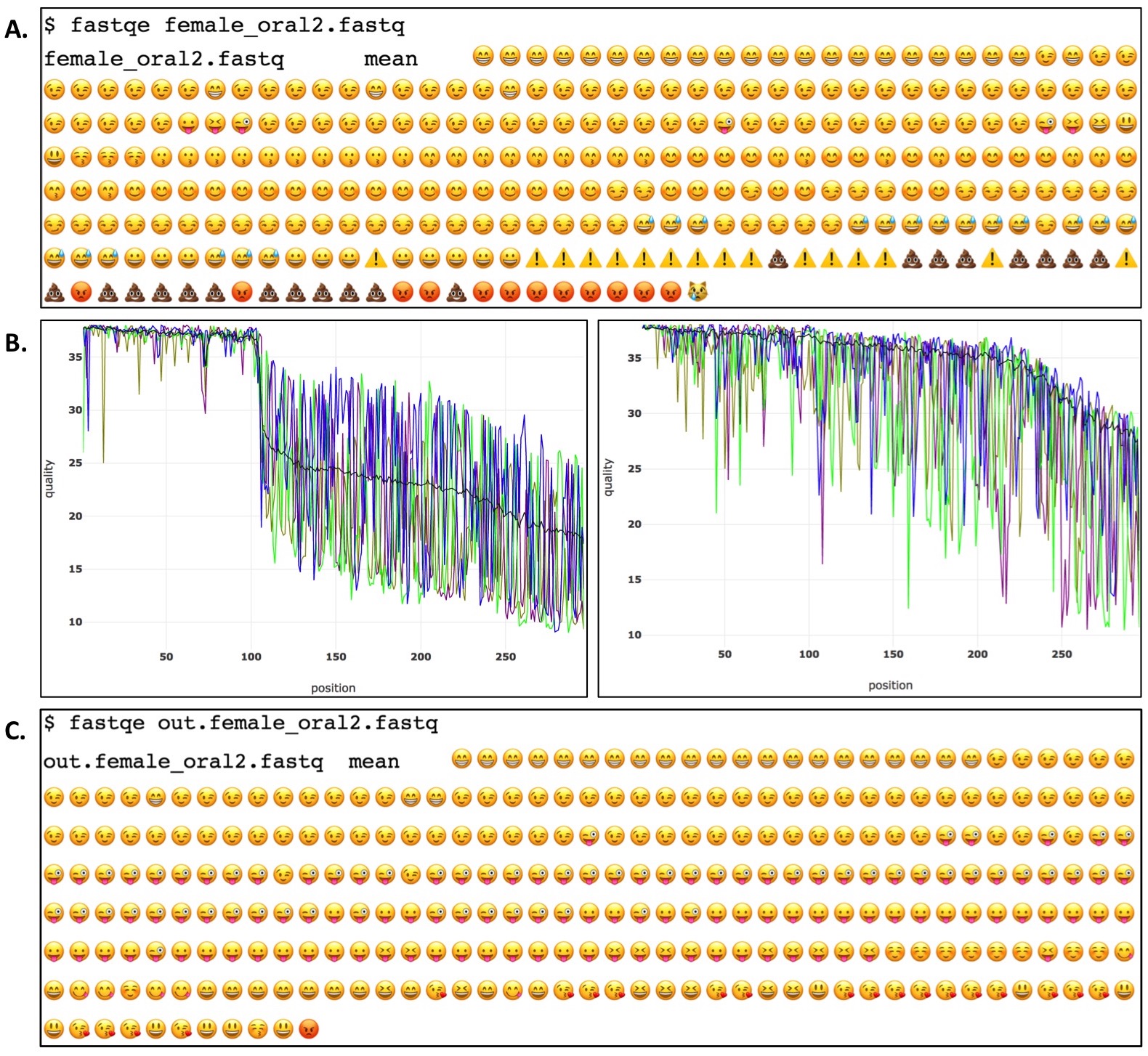
Figure 2. Example of a FASTQE-FASTP genomics command line analysis pipeline. Per base quality of the female_oral2.fastq file is first visualized with FASTQE (A). Low quality bases and sequences are trimmed/filtered from the FASTQ file using the FASTP software package which outputs pre (left) and post (right) adjusted per base quality plots (B). FASTQE is then used again to visualize the trimmed/filtered FASTQ file (C).
Common Mistakes and Sticking Points
For many students, this will be their first experience typing scripts into the command line prompt. Therefore, this activity can be intimidating, particularly when error messages are returned after entering commands. Even though most problems students encounter with this activity are simple, troubleshooting can often be difficult for instructors that are also new to scripting, compounding student frustrations. Though there are many different specific problems students encounter during this activity, Table 2 lists the most common types of mistakes that should be informative for instructors trying to troubleshoot.
Post-class Assessment (optional)
Questions and problems are embedded into the in-class activity. Ideally, these problems are designed for students to work on during the in-class activity; however, instructors may choose to have students work on problems as a take home assignment. Additional extended learning problems are added at the end of the lesson. These problems are recommended for students to work on outside of class either individually or in pairs/groups. Solutions to student questions and problems are available in the instructor version of the lesson (Supporting File S7. FASTQE – Instructor version of lesson).
TEACHING DISCUSSION
Projected estimates for continued exponential growth of the genomics Big Data domain have created an increased need for updating undergraduate life sciences curriculum to include more robust computational bioinformatics training (1). In particular, there is an increased demand for life scientists to develop competencies in computer programming to deal with this data deluge (16-17). Isolated examples of innovative life sciences courses with a programming-based focus have been developed and deployed with successful results (5-10). Unfortunately, these examples reflect the exception rather than the norm in the current landscape of undergraduate life science curriculum. Significant barriers to wide-spread implementation of computational bioinformatics into undergraduate biology courses have limited progress in this area, in particular, lack of faculty training, lack of student interest in bioinformatics, lack of vetted teaching materials, and overly full curricula (4). Alarmingly, this disparity is more prevalent among faculty at minority-serving institutions (MSIs) compared to faculty at non-MSIs (4).
Here we report on the development and integration of a one-off, in-class introductory lesson to genomics command line coding analysis that reinforces several of the NIBLSE core bioinformatics competencies and also surmounts previously reported barriers to curriculum integration (4). Our implementation was in a large enrollment core curriculum genetic course with 278 undergraduate students primarily in the JMU Biology, Biotechnology, and Health Sciences majors. The lesson was specifically designed to reinforce several NIBLSE core competencies including C1. Explain the role of computation and data mining in addressing hypothesis-driven and hypothesis-generating questions within the life sciences, C7. Use command line bioinformatics tools and write simple computer scripts, and C8. Describe and manage biological data types, structure, and reproducibility. (3; https://qubeshub.org/community/groups/niblse/core_competencies).
Of the 183 undergraduate students who completed an optional post course assessment survey, the majority (82.5%) had no substantial training in coding or scripting prior to this lesson. This observation is independent of student's academic level and for the most part academic major demonstrating the general need for increased exposure to scripting in JMU STEM curriculum (Figure 3A, 3D). The outlier group observed in the data is JMU Biotechnology major students who reported higher levels of prior scripting experience (Figure 3D). Only 18 Biotechnology students participated in the survey however, so the validity of this observation will be monitored in future implementations of this lesson. We assessed the effectiveness of achieving our learning outcomes by asking students two additional survey questions at the completion of the lesson. 37.7% of students positively reported that the lesson helped them to better understand complex biological questions (Figure 3B, 3E). Additionally, 31.1% positively reported that the lesson piqued their interest in using command line coding for a future genomics analysis (Figure 3C, 3F). These observations are also independent of student's academic level (Figures 3B, 3C). Students majoring in Biology, Biotechnology, and Health Sciences reported higher levels of understanding and interest than students in majors such as Chemistry, Nursing, Kinesiology, and other non-STEM majors (Figure 3E, 3F). Only 20 students reporting as these "Other" majors participated in the survey however, so the validity of this observation will be monitored in future implementations of this lesson. These data reflect the polarizing nature of exposing STEM students to computer scripting. Representative anecdotal student reflections to the lesson are provided in Table 3.
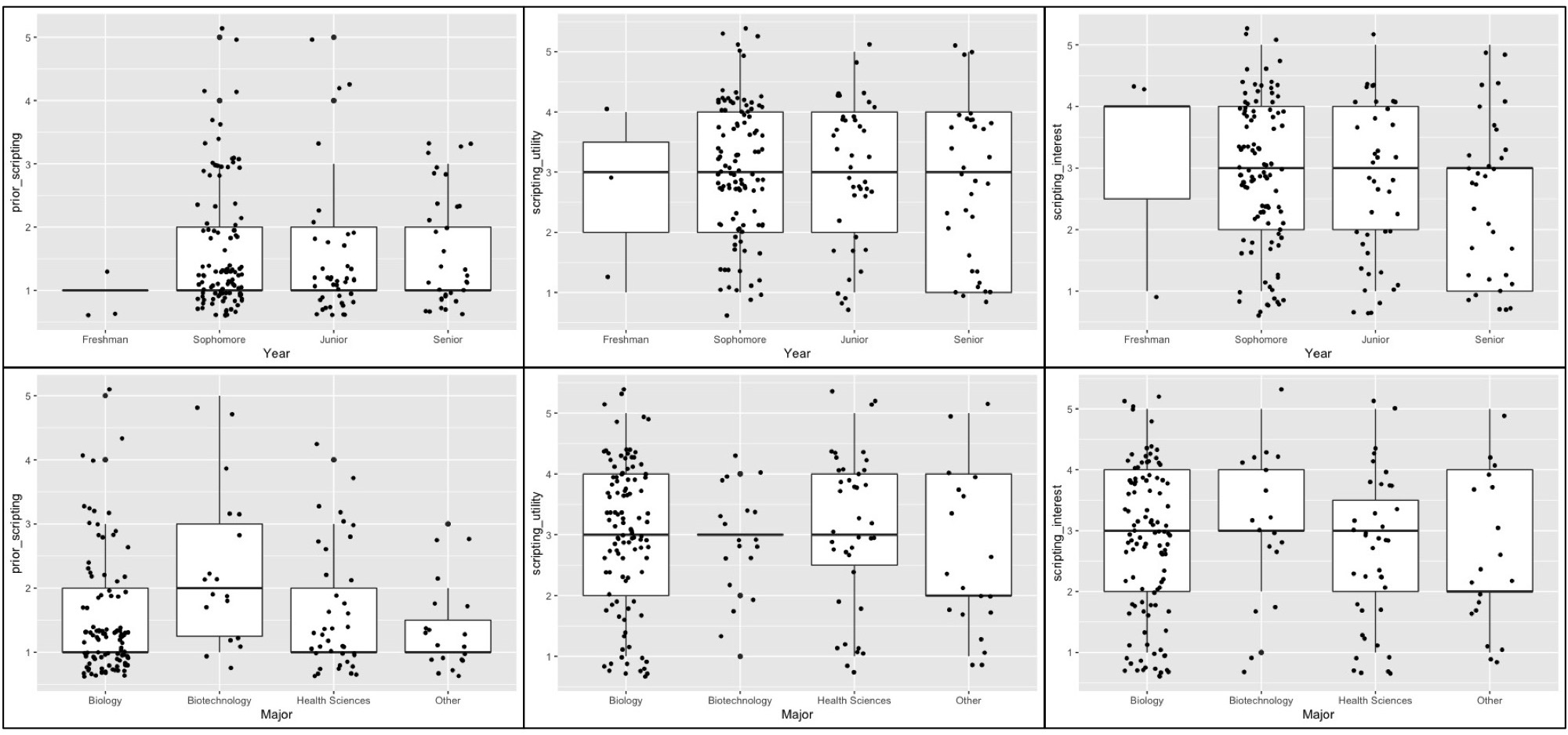
Figure 3. Individually plotted responses of undergraduate student cohort (n = 183) organized by academic year and major to the following post lesson assessment prompts: 1) “Prior to taking this class, I've had adequate training writing simple computer scripts to analyze genomics data (A and D)”; 2) “In class analysis using command line coding helped me to examine and understand complex biological questions.” (B and E); 3) “This lesson has made me more interested in conducting future analyses using command line coding.” (C and F). Student responses were measured using a 5-point Likert scale ranging from 1-“strongly disagree to 5-“strongly agree” and were plotted in RStudio using the ggplot2 package.
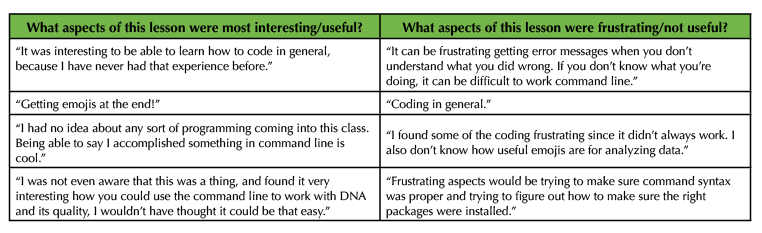
Table 3. Anecdotal student feedback on Introduction to Command Line Programming lesson.
Technical Limitations & Alternative Implementations
A limitation of this lesson is that Windows operating systems do not support the use of emoji symbols; therefore, the FASTQE portion of the command line activity only works on computers running Mac OSX or Linux. To circumvent this technical issue, our team has developed a Jupyter Notebook alternative implementation of this lesson available through the CyVerse cyberinfrastructure platform. This version of the lesson requires setting up a free CyVerse account and launching a Jupyter Notebook web application through CyVerse using step by step instructions which will add approximately 15-20 minutes to the time needed to complete the activity (Supporting File S6. FASTQE – Jupyter Notebook alternative implementation instructions). The upside to this implementation is that instructors and students can run the FASTQE and FASTP portions of the activity from any machine with identical results. Additionally, this implementation is particularly amenable to virtual instruction where instructors have no control over what type of machines will be used.
Several other alternative implementations may also be effectively used. Instructors may choose to demonstrate the FASTQE portion of the lesson to students on a Mac or Linux machine prior to students completing the remainder of the lesson using FASTP QC and filtering software, which is machine agnostic. Alternatively, instructors can substitute the FASTQE portion of the lesson with another widely used command line software such as FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). This lesson works particularly well in classrooms equipped with Anaconda-installed Mac machines. Students can effectively work in pairs or groups of 2-4 on this lesson alternating turns at the keyboard to cut back on the number of machines required. In this situation, providing each student with a different FASTQ file to analyze would sufficiently give each group member a significant portion of the lesson project to manage. Alternatively, this lesson can be effectively implemented into online or hybrid in person/online courses using the Jupyter Notebook implementation maximizing flexibility for both students and instructors (Supporting File S6. FASTQE – Jupyter Notebook alternative implementation instructions). As part of this lesson, we have provided three FASTQ files from an in-class microbiome Illumina sequencing project investigating sexually dimorphic metagenomes associated with garter snake tissues (Supporting File S2. FASTQE – Male5-oral1.fastq file, Supporting File S3. FASTQE – Male5-oral2.fastq file, and Supporting File S4. FASTQE – Female2-oral1.fastq file). However, any FASTQ-formatted data files can be easily substituted into this lesson providing an opportunity for instructors to link learning outcomes to previously established course topics and/or research project. For more advanced students with some coding experience, this lesson would be an effective take home assignment to reinforce basic principles of command line coding.
Our assessment data suggests that this lesson can be an effective early exposure to genomics command line coding (Figure 3; Table 3). Given that the lesson requires minimal or no prior coding experience of both students as well as instructors and that the lesson can be fit into a single class period, several of the most commonly reported barriers to implementing computational bioinformatics into undergraduate curriculum are circumvented (4). Additionally, the novelty of using Emoji data outputs gives students an entertaining hook to help tackle the commonly reported barrier that life science undergraduates find bioinformatics analysis boring or uninteresting. Notably, 85% of upper level undergraduate students (juniors and seniors) in our implementation reported that this was their first exposure to command line analysis (Figure 3A). These data suggest an opportunity to use implementation of our lesson into large-enrollment core curriculum life sciences courses to achieve much broader impact earlier in undergraduate student's academic careers.
SUPPORTING MATERIALS
- S1. FASTQE – Pre-class assignment
- S2. FASTQE – Male5-oral1.fastq file (included in Zip file with all fastq files: S2, S3, and S4)
- S3. FASTQE – Male5-oral2.fastq file (included in Zip file with all fastq files: S2, S3, and S4)
- S4. FASTQE – Female2-oral1.fastq file (included in Zip file with all fastq files: S2, S3, and S4)
- S5. FASTQE – Lecture slides
- S6. FASTQE – Jupyter Notebook alternative implementation instructions
- S7. FASTQE – Instructor version of lesson
- S8. FASTQE – Student version of lesson
ACKNOWLEDGMENTS
The authors would like to thank students in R.A.E.'s BIO481/581 Genomics course for helping to develop this lesson. The authors also thank all BIO240 students and instructors who participated in this study as well as Yasmeen Shorish and Guoqing Lu for providing comments on the manuscript, and M. Rockwell Parker for assisting in sample collection for the in-class garter snake metagenomics project. This work was supported by James Madison University 4-VA funding as well as National Science Foundation, Improving Undergraduate STEM Education Grant #1821657 awarded to R.A.E and the JMU College of Science and Mathematics. All student surveys were conducted with prior approval from the JMU Institutional Review Board for Human Subjects of IRB protocol #17-0307.
References
- Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. 2015. Big data: Astronomical or genomical? PLOS Biology. 12(7), e1002195-e1002195. https://doi.org/10.1371/journal.pbio.1002195
- Levine AG. 2014. An explosion of bioinformatics careers. Science. 344(6189), 1303-1306. https://doi.org/10.1126/science.344.6189.1303
- Sayres MAW, Hauser C, Sierk M, Robic S, Rosenwald AG, Smith TM, Triplett EW, Williams JJ, Dinsdale E, Morgan W, Burnette JM, Donovan SS, Drew JC, Elgin SCR, Fowlks ER, Galindo-Gonzalez S, Goodman AL, Grandgenett NF, Goller CC, Jungck J, Newman JD, Pearson W, Ryder E, Tosado-Acevedo R, Tapprich W, Tobin TC, Toro-Martínez A, Welch LR, Wright R, Ebenbach D, Olney KC, Mcwilliams M, Pauley MA. 2017. Bioinformatics core competencies for undergraduate life sciences education. Cold Spring Harbor Laboratory. https://doi.org/10.1101/170993
- Williams JJ, Drew JC, Galindo-Gonzalez S, Robic S, Dinsdale E, Morgan W, Triplett EW, Burnette J, Donovan S, Elgin S, Fowlks ER, Goodman AL, Grandgenett NF, Goller C, Hauser C, Jungck JR, Newman JD, Pearson W, Ryder E, Sayres MAW, Sierk M, Smith T, Tosado-Acevedo R, Tapprich W, Tobin TC, Toro A, Welch L, Wright R, Ebenbach D, Mcwilliams M, Rosenwald AG, Pauley MA. 2017. Barriers to integration of bioinformatics into undergraduate life sciences education. PloS One, 14(11), e0224288-e0224288. https://doi.org/10.1371/journal.pone.0224288
- Rubinstein A, Chor B. 2014. Computational thinking in life science education. PLoS Computational Biology 10(11), e1003897-e1003897. https://doi.org/10.1371/journal.pcbi.1003897
- Zhan YA, Wray CG, Namburi S, Glantz ST, Laubenbacher R, Chuang JH. 2019. Fostering bioinformatics education through skill development of professors: Big genomic data skills training for professors. PLOS Computational Biology 15(6), e1007026-e1007026. https://doi.org/10.1371/journal.pcbi.1007026
- Tartaro A, Chosed RJ. 2015. Computer scientists at the biology lab bench. Proceedings of the 46th ACM Technical Symposium on Computer Science Education. Paper presented at the 120-125. https://doi.org/10.1145/2676723.2677246
- Mariano D, Martins P, Santos LH, Melo- Minardi RCD. 2019. Introducing programming skills for life science students. Biochemistry and Molecular Biology Education 47(3), 288-295. https://doi.org/10.1002/bmb.21230
- Libeskind-Hadas R, Bush E. 2013. A first course in computing with applications to biology. Briefings in Bioinformatics 14(5), 610-617. https://doi.org/10.1093/bib/bbt005
- Madlung A. 2018. Assessing an effective undergraduate module teaching applied bioinformatics to biology students. PLOS Computational Biology. 14(1), e1005872-e1005872. https://doi.org/10.1371/journal.pcbi.1005872
- Hyman O, Doyle E, Harsh J, Mott J, Pesce A, Rasoul B, Seifert K, Enke RA. 2019. CURE-all: Large scale implementation of authentic DNA barcoding research into first-year biology curriculum. CourseSource 6. https://doi.org/10.24918/cs.2019.10
- Berndsen CE, Young BH, Mccormick QJ, Enke RA. 2016. Connecting common genetic polymorphisms to protein function: A modular project sequence for lecture or lab. Biochemistry and Molecular Biology Education 44(6), 526-536. https://doi.org/10.1002/bmb.20976
- Corpas M, Jimenez RC, Bongcam-Rudloff E, Budd A, Brazas MD, Fernandes PL, Gaeta B, Gelder CV, Korpelainen E, Lewitter F, Mcgrath A, Maclean D, Palagi PM, Rother K, Taylor J, Via A, Watson M, Schneider MV, Attwood TK. 2014. The GOBLET training portal: a global repository of bioinformatics training materials, courses and trainers. Bioinformatics 31(1), 140-142. https://doi.org/10.1093/bioinformatics/btu601
- Donovan S, Eaton CD, Gower ST, Jenkins KP, Lamar MD, Poli D, Sheehy R, Wojdak JM. 2015. QUBES: a community focused on supporting teaching and learning in quantitative biology. Letters in Biomathematics 2(1), 46-55. https://doi.org/10.1080/23737867.2015.1049969
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. Cold Spring Harbor Laboratory. https://doi.org/10.1101/274100
- Stevens SLR, Kuzak M, Martinez C, Moser A, Bleeker P, Galland M. 2018. Building a local community of practice in scientific programming for life scientists. PLOS Biology 16(11), e2005561-e2005561. https://doi.org/10.1371/journal.pbio.2005561
- Baker M. 2017. Scientific computing: Code alert. Nature 541(7638), 563-565. https://doi.org/10.1038/nj7638-563a
- Jacques R, Maza M, Robertson S, Lonsdale A, Enke RA. 2019. A fun introductory command line exercise: Next generation sequencing quality analysis with Emoji! QUBES_FASTQE. QUBES.
Article Files
Login to access supporting documents
A Fun Introductory Command Line Lesson: Next Generation Sequencing Quality Analysis with Emoji!(PDF | 2 MB)
S1.FASTQE-Pre-Class Assignment.pdf(PDF | 4 MB)
S2-4.FASTQE-fastq files.zip(ZIP | 8 MB)
S5. FASTQE-Lecture slides.pptx(PPTX | 12 MB)
S6. FASTQE-Jupyter notebook alternative implementation instructions.docx(DOCX | 1 MB)
S7. FASTQE-instructor version of lesson.pdf(PDF | 1 MB)
S8. FASTQE-student version.pdf(PDF | 888 KB)
- License terms

Comments
Comments
0 Like
Bárbara Bitarello @ on
Using cyverse, the wget command doesn't work because it won't authorize the download. Please assist.
Copy link Report abuse
Comment removed by original commenter @ on (Edited: @ on )