Bioinformatics is a BLAST: Engaging First-Year Biology Students on Campus Biodiversity Using DNA Barcoding
Editor: Srebrenka Robic
Published online:
Abstract
In order to introduce students to the concept of molecular diversity, we developed a short, engaging online lesson using basic bioinformatics techniques. Students were introduced to basic bioinformatics while learning about local on-campus species diversity by 1) identifying species based on a given sequence (performing Basic Local Alignment Search Tool [BLAST] analysis) and 2) researching and documenting the natural history of each species identified in a concise write-up. To assess the student’s perception of this lesson, we surveyed students using a Likert scale and asking them to elaborate in written reflection on this activity. When combined, student responses indicated that 94% of students agreed this lesson helped them understand DNA barcoding and how it is used to identify species. The majority of students, 89.5%, reported they enjoyed the lesson and mainly provided positive feedback, including “It really opened my eyes to different species on campus by looking at DNA sequences”, “I loved searching information and discovering all this new information from a DNA sequence”, and finally, “the database was fun to navigate and identifying species felt like a cool puzzle.” Our results indicate this lesson both engaged and informed students on the use of DNA barcoding as a tool to identify local species biodiversity.
Primary Image: DNA Barcoded Specimens. Crane fly, dragonfly, ant, and spider identified using DNA barcoding.
Citation
Unger S, Rollins M. 2022. Bioinformatics is a BLAST: Engaging First-Year Biology Students on Campus Biodiversity Using DNA Barcoding. CourseSource 9. https://doi.org/10.24918/cs.2022.32Lesson Learning Goals
- Students will know how to use DNA barcoding as a tool to identify species using DNA sequences and understand genetic information.
- Students will be introduced to the NCBI Blast website for use in bioinformatics.
- Students will be exposed to organismal and taxonomic biodiversity to understand how biologist employ the scientific discovery process.
- Students will be engaged and understand the use of DNA barcoding as a tool to identify biodiversity and make connections between organismal and molecular biology.
Lesson Learning Objectives
- Students will be able to demonstrate how to use bioinformatic tools to identify species by performing a BLAST search.
- Students will be able to explain how biologists identify and differentiate species when estimating local biodiversity.
- Students will be able to define DNA barcoding as a technique and evaluate how it relates to organismal biology.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
Science education strives to increase science literacy while informing students across a wide breadth of knowledge and content in an engaging manner (1, 2). The importance of increased training across academic levels is critical for successful careers in STEM fields and success in university (3). Furthermore, actively conducting research can increase student engagement and retention in science (4). However, there are often limitations related to conducting research and bringing research into the classroom (e.g., access to funding, time allotted to more than a small subset of students, involving students in research). Thus, first-year students performing class activities that promote concepts from both fundamental biological concepts, traditionally considered either molecular (e.g., DNA sequencing, PCR, DNA extraction, etc.) or organismal (e.g., biodiversity, taxonomy, etc.) biology, can gain a more holistic approach to the broad field of the biological sciences. Lessons such as the one described here, which help students make connections using tools for identifying species centered on taxonomy, may also help to impart fundamental skills to promote an appreciation of biological diversity and conservation (5) and engage students on local species biodiversity. Measurements of undergraduate attitudes about biodiversity suggest they have a low level of confidence in the ability of science and technology to solve problems resulting from loss of biodiversity (6), thus making incorporation of methods for identification of taxa important in lesson planning at both the high school and college level.
At the undergraduate level, biology lectures and labs have seen an increase in online coursework (7) and remote learning in general. Recently, colleges and universities have seen an increase in online presence, as part of a standard course curriculum (8). Therefore, it is all the more important to develop online lessons which maintain student interest, inform and teach relevant skills to first-year biology students. Educators in biology may struggle to integrate online virtual labs into their courses (9); but ultimately, developing attractive alternatives to in-person laboratories for the online environment should be a goal across biology educators and faculty (10). Specifically, teaching genetics and bioinformatics to biology undergraduates can present additional challenges, either due to the complexity of the material or the lack of fundamental background knowledge retained from the high school science curriculum and courses (11).
DNA barcoding, as a molecular technique has been shown to increase student knowledge of biodiversity loss and provide experience with valuable laboratory skills in DNA extraction and PCR (12). Additionally, DNA barcoding has been used to involve high school students in discovery-based science (13, 14), immerse students in the exciting process of science and modern biological research (15, 16), and promote the notion that DNA is not an abstract idea discussed only in lecture but derives from actual organisms (17). Recently, engaging methods even include BarcodingGO, an analog of the Pokémon GO game (18).
In some cases, universities with sufficient resources are able to include comprehensive lessons, such as a 14-week series of laboratories devoted to DNA barcoding across years, with success in identifying over 1,000 plant, fungi, and invertebrates (19), conduct more rigorous field collections using insect traps and sampling (20), or even 3-week laboratory exercises to explore DNA barcoding (21). Unfortunately, many schools lack the facilities or funds to implement the laboratory portion of extracting DNA and performing PCR. Many investigators readily send samples to more extensive genomic facilities, where services include DNA extraction and sequencing at an ever-decreasing cost to researchers and educators using next-generation sequencing (NGS) technologies (22-24). Moreover, DNA reference libraries, e.g., Barcode of Life Data System, or BOLD (25), provide freely available reference sequences to identify taxa. The Basic Local Alignment Search Tool, or BLAST, is a one-stop platform that allows the easy uploading of DNA sequences using an intuitive user interface (26). This tool enables researchers, educators, and students to upload or paste DNA sequences, then query them (or perform a “BLAST”), and then compare those sequences with a known database for identification.
In this lesson, our goals were essentially three-fold, to develop a concise online lesson that would 1) engage and teach first-year undergraduate biology majors about DNA barcoding using DNA nucleotide sequences to identify species, 2) inform these same students about the local on-campus biodiversity across taxonomic groups, 3) allow students to develop an understanding of the connectivity between molecular and organismal biology and the importance of utilizing both to address fundamental questions. Subsequently, we were also interested in student perceptions on how well this lesson allowed them to perform actual bioinformatics and identify a series of “mystery” species collected from the university’s campus.
Intended Audience
This activity has been implemented in an undergraduate first year level organismal introductory course for biology majors, but it could be incorporated into upper-level biology or ecology courses, especially if sample collection is undertaken by students.
Required Learning Time
A single two- or three-hour laboratory or equal take home time is the minimal time necessary, yet likely varies depending on how much exposure students have had to this topic or their ability to navigate online tools like the BLAST website. For details see Table 1.
Table 1. Lesson Timeline. The lesson spans one full online laboratory but can be modified if delivered in person or if field collection is part of initial lab or if modified to include group or partner reflection in lecture.
| Activity | Description | Estimated Time | Notes |
|---|---|---|---|
| Read 2 articles on DNA barcoding | Pick 2 short general articles on DNA barcoding | ~30 minutes |
We selected two short articles on DNA barcoding to allow students to have some idea of how DNA barcoding is used by biologists to identify species. Articles: |
| Navigate NCBI website | BLAST each DNA barcode sequence | 1 hour | Time may vary based on how long it takes students to follow directions and run through first sequence. Subsequent sequence “blasts” will likely take less than initial run. |
| Research Species identified | Obtain natural history and biological information for each species identified | 1 hour | Students will likely obtain from various online or textbook resources and sources. May vary depending on if invertebrates or vertebrates are used and if covered in lecture. Also depends on level of taxonomic resolution (if identification is down to species or genus or family, etc.). |
| Post Lesson Reflection Questions | Answer reflection question to assess if they liked lesson | 5 minutes | We recommend giving space for students to answer follow up questions in addition to standard Likert scale questions on lesson. |
Prerequisite Student Knowledge
As a background for this lesson, students should be first exposed to basic ideas related to the concepts of speciation, taxonomy, and some level of molecular biology including DNA, sequence composition of DNA, potentially how phylogenetic trees are related to speciation, and what a species is.
Prerequisite Teacher Knowledge
There are no specific teacher prerequisites for instructors with a general knowledge of biology. This lesson can readily be incorporated in both a cellular/molecular course or an organismal course for first year students. However, we recommend instructors go through the lesson and ensure that links are still actively working and also allow plenty of time to receive DNA barcodes ahead of implementation of the lesson. We recommend having a preliminary backlog of several samples run to ensure the activity can be incorporated within a semester, or if done towards the end of a semester, as barcode return time may vary or if students collect samples.
Scientific Teaching Themes
Active Learning
Students will actively use the BLAST website to align DNA barcode sequences to hits in the database (provided by instructors as “mystery DNA”) in order to identify the species of interest. In doing so, they will engage with actual nucleotide sequences, including determining the sequence length, and obtaining the percent identity (how close a match for a specific sequence). Moreover, they will research the species, which includes determining natural history information, biological facts, habitats, common name and other relevant information for each sequence, hence making connections between molecular sequences and species.
Assessment
Students successfully complete a table on correct identification of DNA barcodes to the lowest taxonomic level (typically down to both genus and species), determine sequence length and percent identify, then conduct research on successfully identified species. Therefore, they should have to complete the table before researching correctly the natural history and biological information for each species utilized in this lesson. Students are assessed by if they followed directions and provided correct taxonomic information and adequate natural history information. Students then answer a series of reflection questions to assess whether they thought the activity was helpful in learning about DNA barcoding, and its uses, and how it relates to the organisms, part of their local campus biodiversity. This lesson was provided on Canvas as an online lesson for laboratory, or as a lab.
Inclusive Teaching
This lesson was developed largely as a stand-alone online activity. However, if enough sequences are obtained, this lesson could be a prelude to a further lesson, where students compare what they learned about the species identified. Instructors implementing this activity can incorporate group learning by having students work in small groups of three to four individuals, where this activity can be part of either a laboratory or in-class activity. Groups need access to a device that will allow them to access the BLAST website at NCBI and explore the identified species for natural history traits. The group activity allows students to receive assistance from both the instructor and student peers. Moreover, this computational modeling group activity can appeal to non-traditional learners as it focuses on improving science processing skills, helping students make connections between molecular biology (nucleotide sequences) and organismal biology (species identified). Alternatively, if this activity is provided to students online or as a typical take-home class assignment, students with disabilities can extend the time needed for practice, as this activity can be self-paced. We recommend educators initially demonstrate how to upload sequences subjected to a BLAST, as it may help to guide students that may struggle with technology. This activity is highly customizable to educators. Students can view materials using multiple forms of presentation (with image, without image, using and interpreting simulation results, etc.), thus ultimately increasing the inclusivity of this activity.
Lesson Plan
Overview
A simple short online lesson was developed to engage undergraduate students in a first-year organismal biology course taught asynchronously during COVID-19 pandemic in 2021. At the beginning of this activity, students were given two short articles on DNA barcoding to introduce them to how DNA barcoding using mitochondrial cytochrome c oxidase I gene (CO1) has been used to identify the fish species sold in fish markets and to identify a brown bear from a cave (Table 1). This activity can be prefaced by other articles that could be more directly applicable to how instructors plan on implementing bar-coding. Students were then given “unknown” DNA barcode nucleotide sequences (instructor known sequences of previously submitted samples), from DNA which had been extracted from samples of interest by LifeScanner at ~$10 US per sample following Unger et al., 2020 (27). Students were then asked to identify each “unknown” or “mystery” species using just these sequences (the DNA barcode) down to the lowest taxonomic level, typically the species level. The lesson included detailed instructions for the step-by-step uploading of sequences and performing BLAST (for details, see S1. DNA Barcode Assignment: Mystery Samples). Instructors had previously collected specimens for DNA barcoding during outdoor laboratories by sampling the leaf litter and a small campus pond, which included crayfish, dragonfly, spider, ant, fly, and snail species. DNA was then extracted and sequenced, then sequence length and percent identity of the most likely identification was recorded. Students then were asked to research the species or taxonomic groups identified by collecting information related to the natural history, distribution, and role of each species in the ecosystem. Students then compiled this information into a short paragraph for the six species across various taxonomic groups and also considered why the sequence wasn’t 100% a match.
In order to assess the utility of this activity and our primary research and pedagogy questions, this activity included a short list of survey questions including “Did you enjoy identifying species from on campus?” as a yes or no response. In addition, we asked students to provide feedback on the activity with a short reflection question “Elaborate on whether you enjoyed this activity in at least 2-3 sentences”. Finally, we used a Likert scale to assess understanding of DNA barcoding as a general technique and as a useful tool to identify species by including the following questions “This activity helped me understand DNA barcoding.” And “This activity helped me understand how DNA barcoding is used to identify different species”.
Ethics
This lesson and reflection survey were performed as part of teaching a first-year organismal biology course at a small liberal arts university. All students voluntarily agreed to have their deidentified responses used in this study. This activity follows protocols developed by the Wingate Biology Undergraduate Research Council.
Standard IRB practices were followed in this study including maintaining the anonymity of student participants, data management and protection, and each student agreeing to have their data analyzed for this study. This research posed minimal risk to student participants during normal educational practices and was exempt from IRB approval with the identity of human subjects lacking any identifiers linked to participants in such a manner that the identity was not readily ascertained according to HHS 45 CFR 46104D.
Data Analysis
We exported all submitted, completed lessons and responses from the Canvas Learning Management System (LMS), then compiled these completed activities across all laboratory sections into one single excel file. Next, the authors renamed responses to remove the section or names potentially associated with each file, and included responses from survey questions (Likert scores and written comments) and coded them into an excel file. Responses of participants for short reflection questions on their perception of the usefulness of this activity and whether they felt it helped them understand and use DNA barcoding to identify were assessed as either responses of “Y” for yes and “N” for no, or by using a standard Likert scale. Scores were as follows: 1 corresponded with “Strongly Disagree,” 2 with “Disagree,” 3 with “Undecided,” 4 with “Agree,” and 5 with “Strongly Agree.” We report the descriptive statistics of percentage, median, mean for scores, and quantitative statistics by conducting a chi-squared test on Likert responses using a significance value of 0.05 and combining agree responses and comparing them to disagree responses after removing neutral responses (“undecided”).
Results
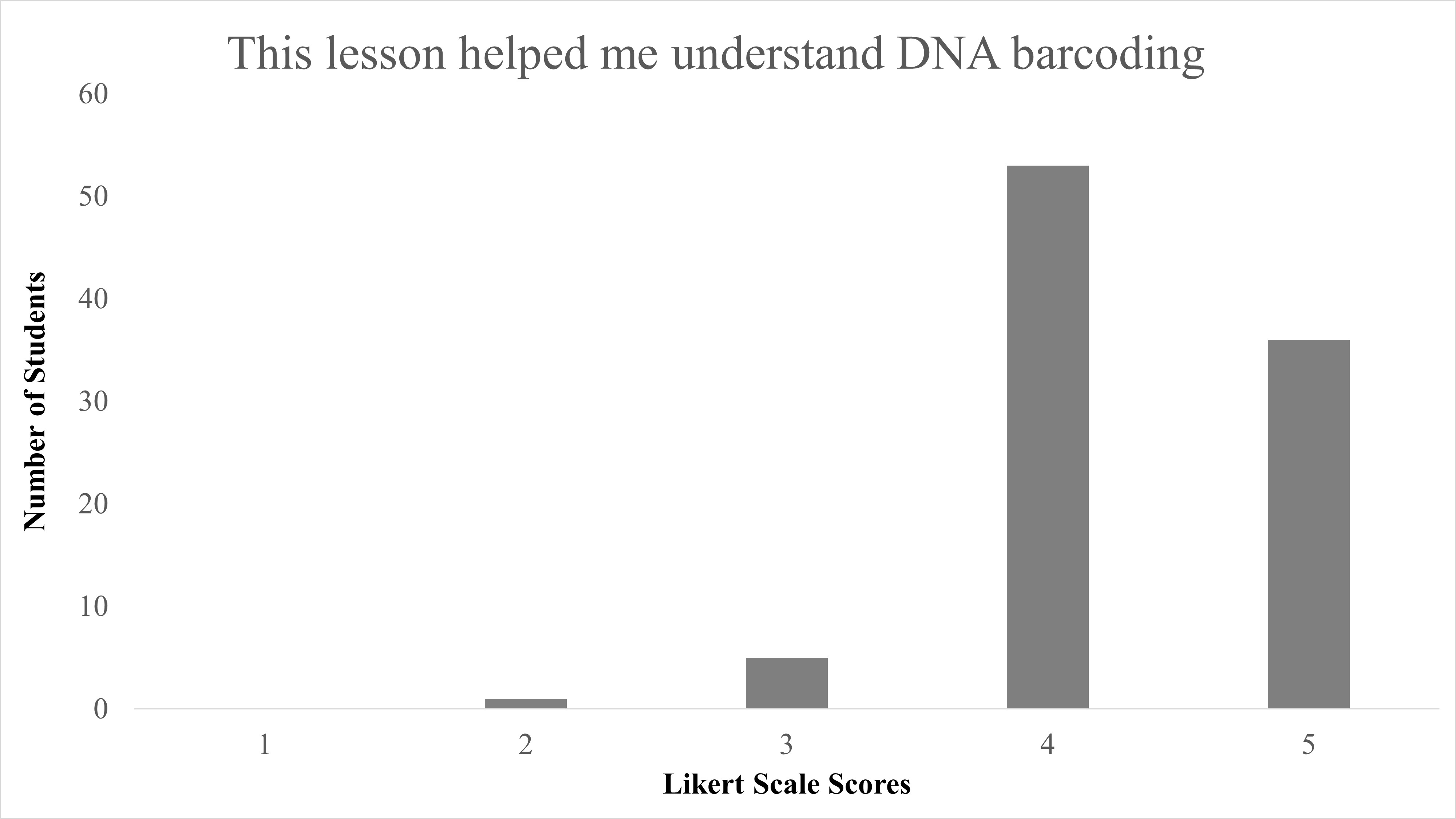
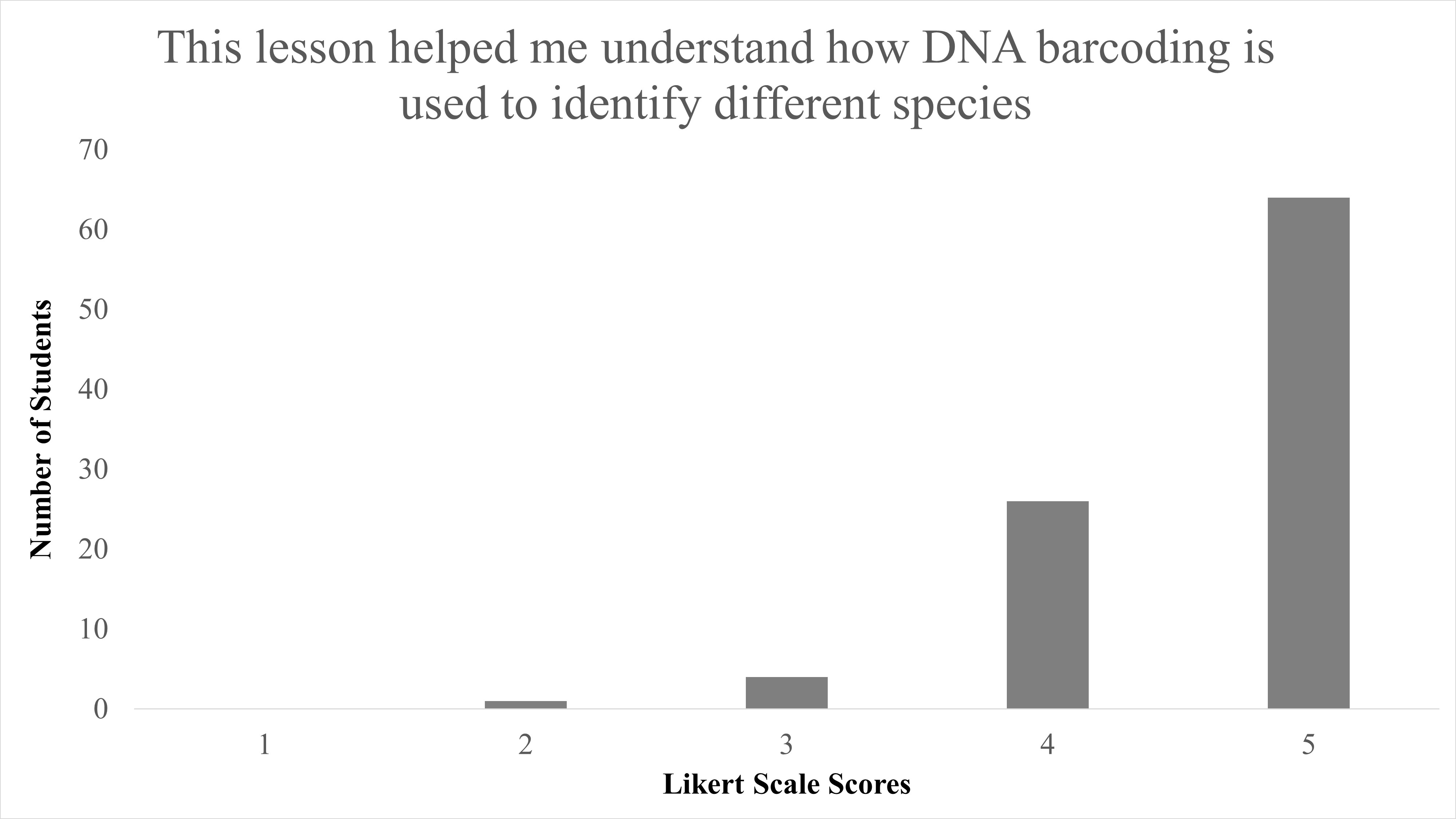
In total, we reviewed 95 lessons turned in by student participants. Responses to the question of “Did you enjoy identifying species from campus?” 89.5% (85/95) answering “Yes,” with only 10.5% (10/95) answering “No” (Table 2). When Likert survey responses to “This lesson helped me understand DNA barcoding” were tabulated and combined, as both “Agree” (53/95) and “Strongly Agree” (36/95), this resulted in 93.68% of student responses agreeing that this activity helped them understand the concept of DNA barcoding and was largely positive (Figure 1). We found a significant difference between responses for disagree vs. agree for the question “This lesson helped me understand DNA barcoding” with X2 (1, N = 90) = 56.5, p < 0.001. When responses to “This lesson helped me understand how DNA barcoding is used to identify different species” were combined as both “Agree” and “Strongly Agree,” this resulted in 94.74% of students agree with the statement on this lesson as beneficial to identify species, and were also largely positive (Figure 2). We found a significant difference between responses for disagree vs agree for responses to the question “This lesson helped me understand how DNA barcoding is used to identify different species” with X2 (1, N = 91) = 57.1, p < 0.001. In summary, students agreed that this lesson helped them both understand the technique of DNA barcoding and how it is used to identify species.
Table 2. Representative student responses to survey questions in activity (n = 95).
| Question | Responses |
|---|---|
| Did you enjoy identifying species from campus: Yes or No? | Yes 89.5%; No 10.5% |
| Elaborate on whether you enjoyed this lesson? |
“I loved working on this activity. I loved searching information and discovering all this new information from a DNA sequence.” “I found the database fun to navigate. Identifying the species felt like a cool puzzle.” “I enjoyed this activity because I have never heard of DNA barcoding and liked learning about species and common names.” “I liked learning about what species were on campus, I never thought about the insects.” “This activity shows how a specific DNA sequence is linked to an organism which I enjoyed looking for matches of the DNA sequence.” “This was very enjoyable even though it was online. Being able to find out what an organism is by searching and looking up a nucleotide sequence is very satisfying” |
| This lesson helped me understand DNA barcoding. | Median = 4; Mean = 4.3 ± 0.62 S.D. |
| This lesson helped me understand how DNA barcoding is used to identify species. | Median = 5; Mean = 4.6 ± 0.623 S.D. |
All students successfully identified sequences to larger taxonomic group and common name, with the majority of students reporting down to the lowest taxonomic level of species for Libulla incesta (slaty skimmer dragonfly), Procambarus acutus (White River crayfish), Camponotus pennsylvanicus (black carpenter ant), Latrodectus variolus (northern black widow spider), and Physella acuta (tadpole snail). The sequence length varied from 144 to 613, with an average of 424.5 nucleotides. 100% of students correctly identified species to some taxonomic, with 5/6 organisms identified down to the species level, mostly with high percent identity. However, most students reported one of the identified species down to only the Order or Family level (Dipteran sample) due to the lack of database results, not student error. Interestingly, several students reported that they did not realize that some species were not native, including the tadpole snail identified, which is established across the southeastern US (28).
Representative responses to our questions on elaborate on this lesson (Table 2), included the following: “I loved working on this activity. I loved searching information and discovering all this new information from a DNA sequence, that is something that will never stop amazing me. I never really looked into the animal part of biology because I grew up with the stereotyped idea that it was boring but now, I am having to rethink my career path into biology” as well as “I thought it was interesting that so many possible species could be found on campus” and “I enjoyed this activity because it allowed me to identify species using the method I learned about. Application is one of the main ways I learn, so applying the techniques learned allowed me to really understand.” When taken together, these responses highlight the utility of this activity to successfully engage and educate students using DNA barcoding sequences.
Other students engaged in the BLAST search in a different way noting that “…it was cool to see the different types of organisms come up. I would try to predict what it might be from the scientific name…,” highlighting the anticipation of the experimental process. Others noted, “Analyzing these species on our own allowed us to understand their classification and how they interact with their environment” and “Now I have a better idea of what scientists are doing when they DNA barcode.” Additional responses include “I found the database fun to navigate. Identifying the species felt like a cool puzzle” and “I like being able to make the connection between the DNA and the species and seeing how many different species are closely related to the sequence that was put in.” Written comments were overwhelmingly positive, with student reflections highlighting how this online lesson helped them bridge the gap between molecular (DNA sequences) and organismal biology (local biodiversity). Finally, the majority of students reported natural history information for each species including the geographic range of species identified, habitats, conservation status, feeding habits or diet, as well as ecological information (e.g., species role as predator or prey or if sensitive to pollution, etc.) and overall biology or taxonomy of species.
Teaching Discussion
This lesson provided students with an engaging, quick, self-directed online format for learning about how bioinformatics works and how DNA barcoding can be utilized to identify species to the lowest taxonomic levels. Moreover, students researched and reported information about the natural history of the species they successfully identified. Several students noted in their write-ups on the natural history of one of the identified species, Physella acuta, as being invasive to other areas such as Europe. This highlights not only the potential for students to have increased awareness of local species diversity, but that by researching each animal identified, students were allowed to learn more about specific taxonomic groups. Therefore, when incorporating this lesson, we recommend sampling across multiple taxa as a model for teaching about organismal taxonomy, conservation, and even invasive species. Other researchers have used DNA barcoding as a way to involve students in community or citizen science endeavors with invasive species, such as the lionfish, Pterois volitans (29). Educators can sample commonly established non-native species in their local areas around the campus to use those sequences and subsequent DNA barcode identification as a teaching moment to impart the reality of invasive species issues to undergraduates. We recommend science educators (either high school or college biology instructors) adapt this lesson to their own region.
This lesson also provides a number of advantages to both students and faculty teaching in the life sciences. The kits we utilized, allowing four DNA barcoding samples to be analyzed, are easy and affordable (~$54 USD per kit for 4 samples), and require little to no lab work. If possible, this lesson could be adapted into existing outdoor laboratories by collecting more specimens each semester following collection procedure highlighted in kits, i.e., small insect wing or portion of leg included and sent for sequencing. Integrating DNA barcoding into an undergraduate curriculum can expose students to a wide range of scientific concepts, problem-solving skills, inquiry, and both field and laboratory work (17). Alternatively, if educators have limited funding, they can modify this online lesson using previously published DNA sequences available at no cost on the NCBI BLAST website.
The teaching of this lesson could involve students, individually or in small groups, collecting samples, as this may provide a more hands-on approach for field collection, such as we had performed during outdoor labs before 2020. The incorporation of DNA barcoding can serve as a bridge between upper and lower-level coursework (30). Barcoding kits now even allow for the identification of plants, allowing for greater flexibility to identify species if the course is taught with more of an emphasis on plant biology. We also recommend educators with access to basic genetic laboratories and equipment (PCR machines, gel electrophoresis) to extract the DNA using standard kits and protocols readily available to provide additional opportunities to make connections between laboratory protocols and endpoints of species identification (31). While DNA barcoding has been included as an established research and education tool in upper-level undergraduate biology laboratories (32), not all schools have the access or funding to perform the DNA extractions themselves. Therefore, this lesson provides an alternative for DNA barcoding with affordable kits which have been utilized to identify a variety of taxa (33), with reasonable turn-around times to focus on the bioinformatics of identification to connect actual DNA sequences to the organism which students identify and research.
One caveat of this lesson is that not every species may be identified to its lowest taxonomic level (or species) based on the availability and similarity of reference DNA in the NCBI database. Other studies have found this to be the case, with larval fish species only being identified down to family or genus (34). We noted this for our Diptera, Tipula, where students often did not report down to either Family, Genus, or species level. This lack of specification was usually due to low matching % ID and lack of taxonomic resolution of the database, not due to student error. While this observation may, on the surface, seem like a disadvantage, we consider this to be an additional teaching moment for students to recognize that not all species can currently be identified down to the species level using the database. This can be further followed up with some discussion of species concepts and factors associated with the lack of taxonomic resolution for identification (e.g., limited samples, short sequence length, or lack of representation in database, etc.). We anticipate this may be the case for lesser studied groups across regions which lack reference barcodes, e.g., aquatic invertebrates, marine mollusks, ascidians, etc. (35). We encourage further discussion of reasons why all species or taxa are not equally represented in the database. Educators can even use this result to discuss species concepts in general.
A small subset of surveyed students, ~ 5%, reported they found the navigation of the BLAST website/tool confusing at first but subsequently said it was easier after using it for multiple species. One example student response stated, “this activity was a bit confusing but it was honestly a cool website and I thought it was interesting how it pulled up so much information.” To reduce initial confusion, we recommend that instructors demonstrate how to upload and subject sequences to BLAST analysis before having the students perform this step. An example could be done in person as part of a lecture or laboratory. The instructor could create a short video or broadcast the demonstration over a video conferencing platform for online courses.
To ensure students participating in the activity achieve the outcomes and goals of learning both the concept and application of DNA barcoding, several follow-up assessment exam or quiz questions could be included. These could ensure that students are able to apply information learned to a new scenario. Example questions include:
Q1. Given a mystery sequence of DNA, how would you determine the probable species?
Answer: Open-ended or multiple-choice format, involving a description of navigating to the NCBI BLAST website, copy and pasting sequence, running sequence, and assessing the percent identity as it pertains to the probable species.
Q2. Provide students an output example (4-5 likely choices) from a BLAST which includes possible species and percent identity unsorted or out of order (i.e., percent identity or % likely species randomized in the answer)
Answer: Multiple-choice, with students selecting the correct answer based on the higher percent identity. This could further be emphasized in an activity where the output parameter “Per. Ident.” is clearly defined as “how similar the query sequence is to the target sequence” with the higher probability more likely a match.
Q3. Give students a real or a fictitious output from a BLAST which includes both percent identity and the query length, which corresponds with lower query length, a lower percent identity.
Answer: Open-ended, with students discussing how nucleotide sequence length as a parameter relates to the percent identity when comparing query sequences in the database. Educators can use a real example to generate this question, a sequence length of ~ 500, then deleting specific portions of either the beginning or end of the known sequence down to examples which are either 400, 300, 200, to 100, and develop the question as percent identity lowers according to lower sequence length. Additionally, educators could have students analyze the E value (expected value) in the output of their BLAST, which describes how many times one would expect a match by chance in a database of that particular size (the lower the E value, the more significant the match). This could help students further interpret the probability of a valid/useful match when generating their identifications.
Q4. What are the applications of DNA barcoding?
Answer: Open-ended or multiple-choice. Answers should include assessment of whether expensive products contain the correct fish or meat as advertised, the use of DNA barcoding in conservation biology and biodiversity, and lastly, the potential for DNA barcoding in a variety of ecological studies, such as identifying dietary items from animals, identifying pests or invasive species, or as a way to align molecular and organismal biology.
Conclusion
There is a growing body of research on best pedagogical practices to engage undergraduate biological science majors by incorporating hands-on, active learning methodologies in laboratories and lecture classrooms. Many of these methods are becoming increasingly affordable and are easily accessible to educators with modest school budgets. These activities can provide a transformative experience and serve as a learning bridge between molecular biology, community ecology, and local biodiversity. Our results show that for a modest investment (< $108 for two DNA barcoding kits submitted for sequencing of 8 total species) and without the need to perform extractions or PCR in a laboratory, undergraduate students can become more informed about on-campus biodiversity, learn about several taxonomic groups, all while making the connection between DNA sequences and the individual organism by performing BLASTs.
Supporting Materials
-
S1. DNA Barcode Assignment: Mystery Samples – Example of online lesson assignment
Acknowledgments
We want to thank Organismal Biology students of 2020-2021, the Wingate Biology Department, and the Biology Research Council for allowing us to conduct this study. Several undergraduate laboratory students in Organismal Biology helped us collect organisms for this activity in 2020, so we thank them for their hard work. We also thank David and Myra Seitz for their aid in developing engaging activities in biological science which fostered the importance of science education in author.
References
- Bradforth SE, Miller ER, Dichtel WR, Leibovich AK, Feig AL, Martin JD, Bjorkman KS, Schultz ZD, Smith TL. 2015. University learning: improving undergraduate science education. Nature 532:282–284. doi:10.1038/523282a.
- McComas WF. 2017. Understanding how science works: The nature of science as the foundation for science teaching and learning. Sch Sci Rev 98:71–76.
- Rozek CS. Svoboda RC, Harackiewicz JM, Hulleman CS, Hyde JS. 2017. Utility-value intervention with parents increases students’ STEM preparation and career pursuit. PNAS 114:909–914. doi:10.1073/pnas.1607386114.
- Henter HJ, Imondi R, James K, Steinke D. 2016. DNA barcoding in diverse educational settings: five case studies. Phil Trans R Soc B 371:20150340. doi:10.1098/rstb.2015.0340
- Bilton DT. 2014. What’s in a name? what have taxonomy and systemics ever done for us?. J Biol Educ 48:116–118. doi:10.1080/00219266.2014.926653
- Huang H, Lin Y. 2014. Undergraduate students’ attitudes toward biodiversity. Univers J Educ Res 2:379–386. doi:10.13189/ujer.2014.020406.
- Biel R, Brame CJ. 2016. Traditional versus online biology courses: connecting course design and student learning in an online setting. J Microbiol Biol Educ 17:417–422. doi:10.1128/jmbe.v17i3.1157.
- Korkmaz G, Toraman C. 2020. Are we ready for the post-COVID-19 educational practice? An investigation into what educators think as to online learning. I J Techn Educ Sci 4:293–309. doi:10.46328/ijtes.v4j4.110.
- Wolinsky H. 2021. Biology lessons in times of COVID. EMBO reports 22:e52513. doi:10.15252/embr.202152513.
- Procko K, Bell JK, Benore MA, Booth RE, Moore V, Dries DR, Martin DJ, Mertz PS, Offerdahl EG, Payne MA, Vega QC, Provost JJ. 2020. Moving biochemistry and molecular biology courses online in times of disruption: recommended practices and resources- a collaboration with the faculty community and ASBMB. Biochem Mol Biol Educ 2020:1–7. doi:10.1002/bmb.21354.
- Gibson DJ, Gibson LS. 1993. College students’ perceptions on adequacy of high school science curriculum as preparation for college level biology. Am Biol Teach 55:8–12. doi:10.2307/4449571.
- Florio A, Rivera, N, Nolan K. 2018. The use of DNA barcoding to teach students the importance of classifying biodiversity. Tested Studies Lab. Teach. 39:1–10.
- Butler M, Henter H, Mel S. 2014. From bugs to barcodes: using molecular tools to study biodiversity. Tested Studies Lab. Teach. Proceed. Assoc. Biology Lab. Educ. 35:41–55.
- Santschi L, Hanner RH, Ratnasingham S, Riconscente M, Imondi R. 2013. Barcoding life’s matrix: translating biodiversity genomics into high school settings to enhance life science education. PLOS Biol 11:1–8. doi:10.1371/journal.pbio.1001471.
- Harris S, Bellino M. 2013. DNA barcoding from NYC to Belize. Science 342:1462–1463. doi:10.1126/science.1230006.
- Musante S. 2010. DNA Barcoding investigations bring biology to life. BioScience 60:14. doi:10.1525/bio.2010.60.1.4.
- Shevchenko J, Yuni ABM, Frandsen PB. 2019. Undergraduate teaching of scientific concepts using DNA barcoding of Trichoptera. Zoosymposia 14:16–31. doi:10.11646/zoosymposia.14.1.4.
- Nunes, R, Oliveira I, Dias P, Bidinotto AB, Telles M. 2020. BarcodingGo: A problem-based approach to teach concepts related to environmental-DNA and bioinformatics. Biochem Mol Biol Educ 49:210–215. doi:10.1002/bmb.21424.
- Hyman OJ, Doyle EA, Harsh J, Mott J, Pesce A., Rasoul B, Seifert K, Enke RA. 2019. CURE-all: Large scale implementation of authentic DNA barcoding research into first-year biology curriculum. CourseSource 6. doi:10.24918/cs.2019.10.
- Steinke D, Brenton V, Berzitis E, Herbert PDN. 2017. The school malaise trap program: coupling educational outreach with scientific discovery. PLoS Biol 15:e20011829. doi:10.1371/journal.pbio.2001829.
- Erasmus DJ. 2021. DNA barcoding: a different perspective to introducing undergraduate students to DNA sequence analysis. Biochem Mol Biol Educ in press. doi:10.1002/bmb.21492.
- Alekseyev YO, Fazeli R, Yang S, Basran R, Maher T, Miller NS, Remick D. 2018. A next-generation sequencing primer —how does it work and what can it do?. Acad Pathol 5:1–11. doi:10.1177/2374289518766521.
- Olsen RJ, Long SW, Musser JM. 2012. Bacterial genomics in infectious disease and the clinical pathology laboratory. Arch Pathol Lab Med 136:1414 –1422. doi:10.5858/arpa.2012-0025-RA.
- Slatko BE, Gardner AF, Ausubel FM. 2018. Overview of next generation sequencing technologies. Curr Protoc Mol Biol 122:e59. doi:10.1002/cpmb.59.
- Ratnasingham S, Hebert PDN. 2007. BOLD: the barcode of life data system. (www.barcodinglife.org). Mol Ecol Notes 7:355–364. doi:10.1111/j.1471-8286.2007.01678.x.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acid Res 26:W5–W9. doi:10.1093/nar/gkn201.
- Unger SD, Williams LA, Diaz L, Bodinof-Jachowski C. 2020. DNA barcoding to assess diet of larval eastern hellbenders in North Carolina. Food Webs e00134. doi:10.1016/j.fooweb.2019.e00134.
- Benson A. 2007. Physella acuta. USGS nonindigenous aquatic species database, Gainesville, FL. https://nas.er.usgs.gov.querries.
- Eble J, Pecore J. 2019. “Invasive Aliens”: a student citizen-science activity using DNA barcoding to investigate concepts in ecology and molecular biology. Am Biol Teach 81:169–174. doi:10.1525/abt.2019.81.3.169.
- Melvin P. 2020. Building community: using DNA barcoding through a collaborative project between lower-division and upper-division STEM courses. Essays on Best Practices in the University System of Georgia 2:37–39.
- Naaum AM, Frewin A, Hanner R. 2013. DNA barcoding as an educational tool: case studies in insect biodiversity and seafood identification. Teach Learn Innov J 16:25–33.
- Dodgen CD, Newman L, Lee C. 2017. DNA barcoding, NCBI data tool and Mega as a teaching and research tool for undergraduate biology laboratory activities. GA J Sci 75:21. https://digitalcommons.gaacademy.org/gjs/vol75/iss1/21.
- McKeon S, Fenolio D, Dreelin A, Shaw D, Kobrinsky Z, Meyer C. 2020. Nextgen natural history: new techniques for classical natural history questions. J Nat Hist Educ Exp 14:6–12.
- Chu C, Loh KH, Ng CC, Ooi AL, Konishi Y, Huang S, Chong VC. 2019. Using DNA barcodes to aid in the identification of larval fishes in tropical estuarine waters (Malacca Straits, Malaysia). Zool Stud 58:30. doi:10.6620/ZS.2019.58-30.
- Curry CJ, Gibson JF, Shokralla S, Hajibabaei M, Baird DJ. 2018. Identifying North American freshwater invertebrates using DNA barcodes: are existing COI sequence libraries fit for purpose?. Freshw Sci 37:178–189. doi:10.1086/696613.
Article Files
Login to access supporting documents
Unger-Rollins-Bioinformatics is a BLAST.pdf(PDF | 179 KB)
S1. DNA Barcode Assignment Mystery Samples - Example of online lesson assignment.pdf(PDF | 334 KB)
- License terms

Comments
Comments
There are no comments on this resource.