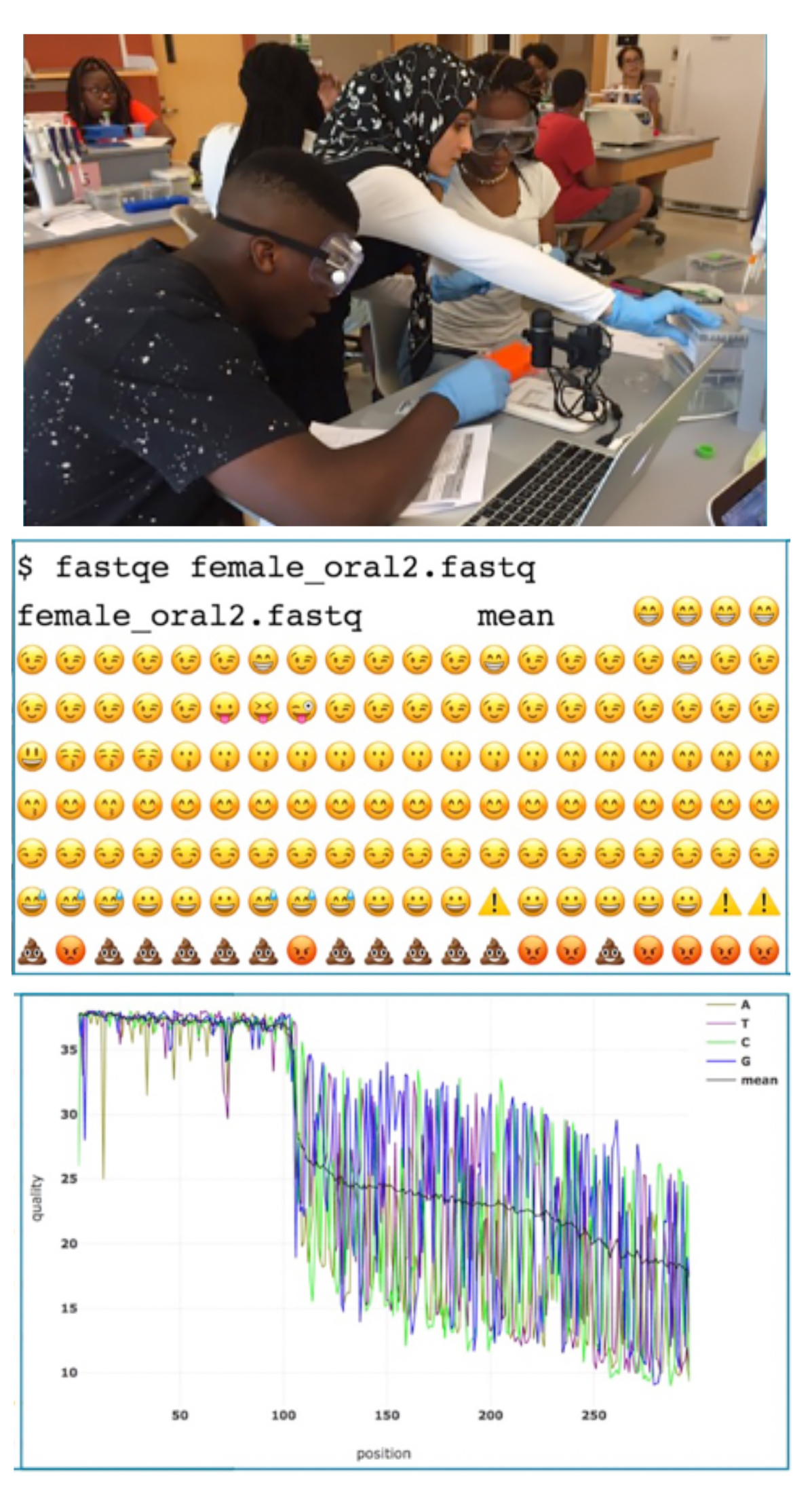
Radical innovations in DNA sequencing technology over the past decade have created an increased need for computational bioinformatics analyses in the 21st century STEM workforce. Recent evidence however demonstrates that there are significant barriers to teaching these skills at the undergraduate level including lack of faculty training, lack of student interest in bioinformatics, lack of vetted teaching materials, and overly full curricula. To this end, the James Madison University, Center for Genome & Metagenome Studies (JMU CGEMS) and other PUI collaborators are devoted to developing and disseminating engaging bioinformatics teaching materials specifically designed for streamlined integration into general undergraduate biology curriculum. Here, we have developed and integrated a fun introductory level lesson to command line next generation sequencing (NGS) analysis into a large enrollment core biology course. This one-off activity takes a crucial but mundane aspect of NGS quality control (QC) analysis and incorporates the use of Emoji data outputs using the software FASTQE to pique student interest. This amusing command line analysis is subsequently paired with a more rigorous research-grade software package called FASTP in which students complete sequence QC and filtering using a few simple commands. Collectively, this short lesson provides novice-level faculty and students an engaging entry point to learning basic genomics command line programming skills as a gateway to more complex and elaborated applications of computational bioinformatics analyses.
Primary image: Undergraduate students learn the basics of command line NGS quality analysis using the FASTQE and FASTP programs.