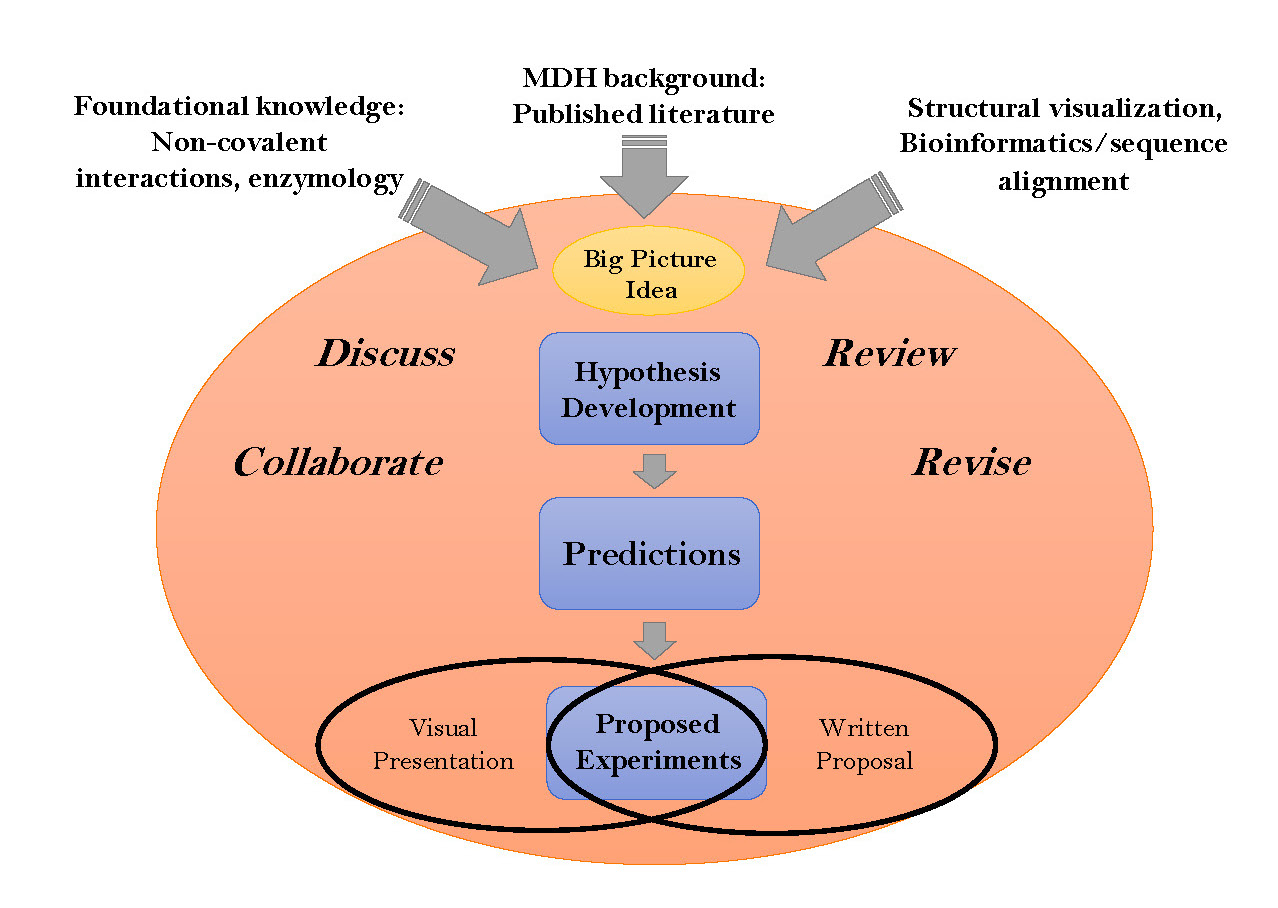
Developing student creativity and ability to develop a testable hypothesis represents a significant challenge in most laboratory courses. This lesson demonstrates how students use facets of molecular evolution and bioinformatics approaches involving protein sequence alignments (Clustal Omega, Uniprot) and 3D structure visualization (Pymol, JMol, Chimera), along with an analysis of pertinent background literature, to construct a novel hypothesis and develop a research proposal to explore their hypothesis. We have used this approach in a variety of institutional contexts (community college, research intensive university and primarily undergraduate institutions, PUIs ) as the first component in a protein-centric course-embedded undergraduate research experience (CURE) sequence. Built around the enzyme malate dehydrogenase, the sequence illustrates a variety of foundational concepts from the learning framework for Biochemistry and Molecular Biology. The lesson has three specific learning goals: i) find, use and present relevant primary literature, protein sequences, structures, and analyses resulting from the use of bioinformatics tools, ii) understand the various roles that non-covalent interactions may play in the structure and function of an enzyme. and iii) create/develop a testable and falsifiable hypothesis and propose appropriate experiments to interrogate the hypothesis. For each learning goal, we have developed specific assessment rubrics. Depending on the needs of the course, this approach builds to an in-class student presentation and/or a written research proposal. The module can be extended over several lecture and lab periods. Furthermore, the module lends itself to additional assessments including oral presentation, research proposal writing and the validated pre-post Experimental Design Ability Test (EDAT). Although presented in the context of course-based research on malate dehydrogenase, the approach and materials presented are readily adaptable to any protein of interest.
Primary image: Mind map of the hypothesis development.

Susan L Klinedinst onto Biochemistry
@
on