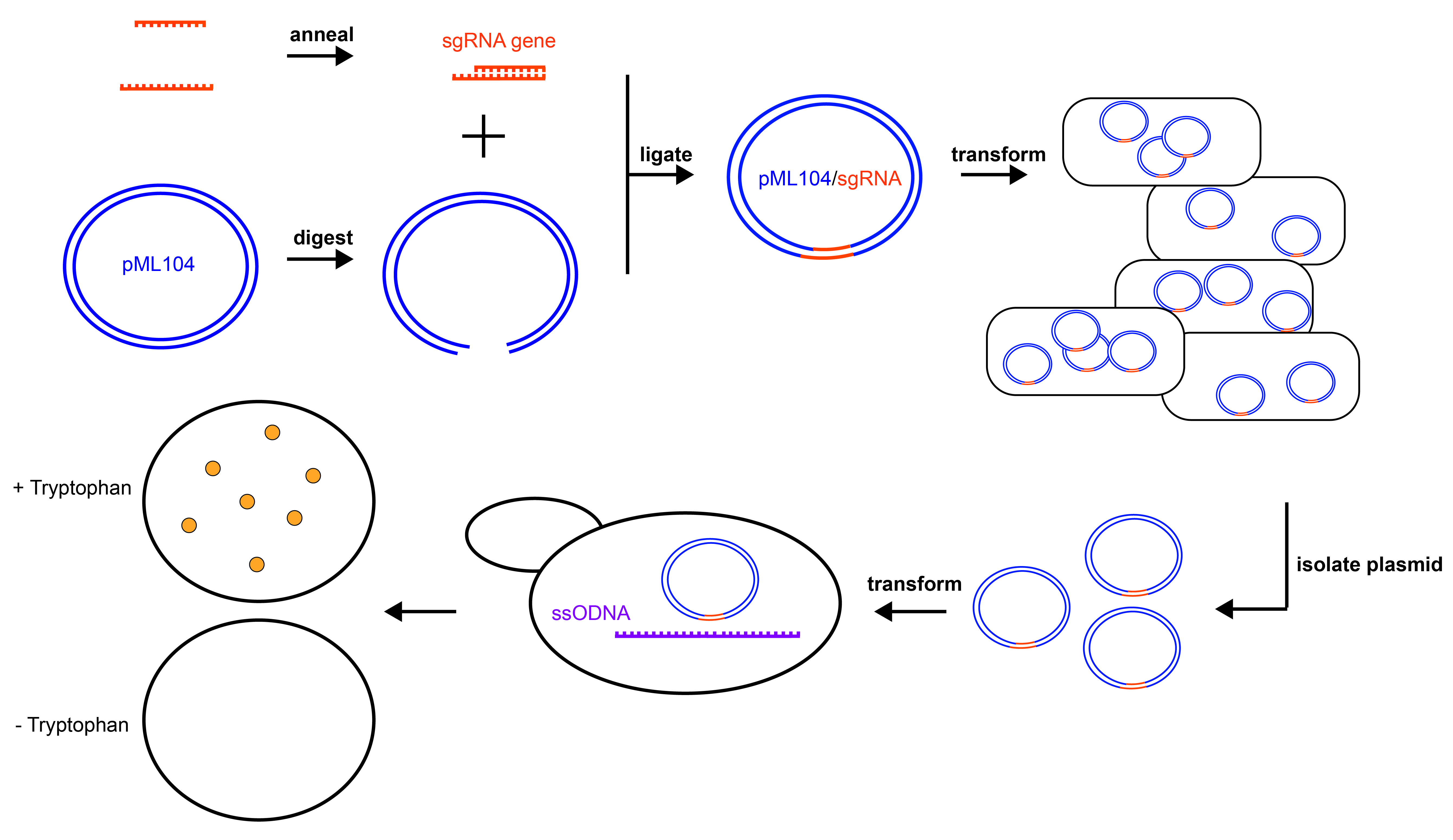
Providing undergraduate life-science students with a course-based research experience that utilizes cutting-edge technology, is tractable for students, and is manageable as an instructor is a challenge. Here, I describe a multi-week lesson plan for a laboratory-based course with the goal of editing the genome of budding yeast, Saccharomyces cerevisiae. Students apply knowledge regarding advanced topics such as: CRISPR/Cas9 gene editing, DNA repair, genetics, and cloning. The lesson requires students to master skills such as bioinformatics analysis, restriction enzyme digestion, ligation, basic microbiology skills, polymerase chain reaction, and plasmid purification. Instructors are led through the technical aspects of the protocols, as well as the teaching philosophy involved throughout the laboratory experience. As it stands, the laboratory lesson is appropriate for 6-8 weeks of an upper-level undergraduate laboratory course, but may be adapted for shorter stints and students with less experience. Students complete the lesson with a more realistic idea of life science research and report significant learning gains. I anticipate this lesson to provide instructors and students in undergraduate programs with a hands-on, discovery-based learning experience that allows students to cultivate skills essential for success in the life sciences.

Carolyn Wetzel onto Genetics BIO243
@
on