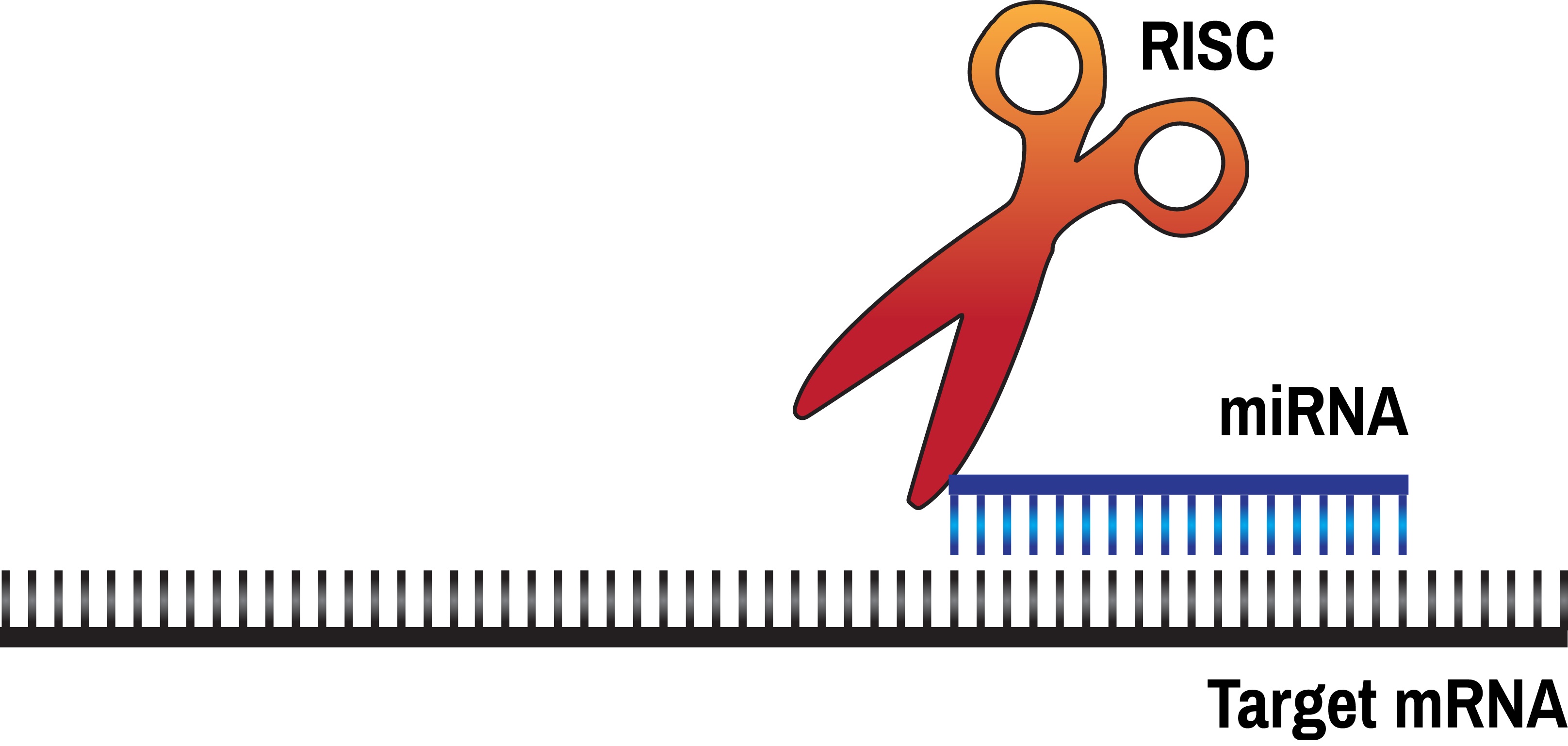
Incorporating authentic research experiences into undergraduate labs, while shown to be particularly effective at engaging and retaining students in STEM majors, can be difficult to accomplish within the constraints of resource availability or cost, and time limitations. One area that is particularly amenable to adaptation for undergraduate lab classes is the discovery and validation of targets of microRNAs (miRs). The human genome encodes several hundred, possibly several thousand miRs, each of which is a 22 nucleotide long RNA molecule capable of regulating the expression of multiple target genes. miRs have been shown to be critical during development, for human health and disease, and are currently being investigated as both therapeutic agents, as well as possible drug targets. A lack in understanding the mechanisms by which miRs recognize their targets makes computer-based predictions of miR targets quite inaccurate, necessitating experimental verification of such predictions. In this lesson, we describe an easily adaptable lab module that can be used in existing undergraduate molecular biology lab courses to conduct authentic scientific research. Students use a variety of databases to identify likely candidate genes whose expression may be altered by a given miR, and then experimentally test their predictions in human cells. This inquiry-based module gives students a taste of real scientific research and excites them about the possibility that, even as a student, they have the potential to contribute to this cutting edge research.

Rob Levenson onto Biochemistry
@
on