The central dogma of biology is a foundational concept that is traditionally included in genetics curricula at all academic levels. Despite its ubiquitous presence throughout genetics education, students persistently struggle with both the fundamental and advanced topics of the central dogma. In particular, students conflate the role of genomic variations in DNA replication, transcription, and translation. As research and healthcare increasingly utilizes genomic medicine to link genetic variants to clinical phenotypes, it is critically important for biology and health science students to understand the role of genetic variation in molecular genetics. This lesson focuses on the role of missense mutations in the central dogma using a case study of cystic fibrosis. The case study is paired with a creative activity in which students draw the molecular parts of the central dogma. This helps students to connect the abstract concepts of the central dogma to a real-world clinical example. The effectiveness of this lesson was evaluated for two semesters of a Human Genetics course using end-of-unit exam questions. The active-learning lesson is an engaging activity that reinforces the role of genetic variation in the central dogma and the effects on clinical phenotypes. This lesson is highly customizable and adaptable to courses of various sizes, levels, course lengths, and teaching modalities.
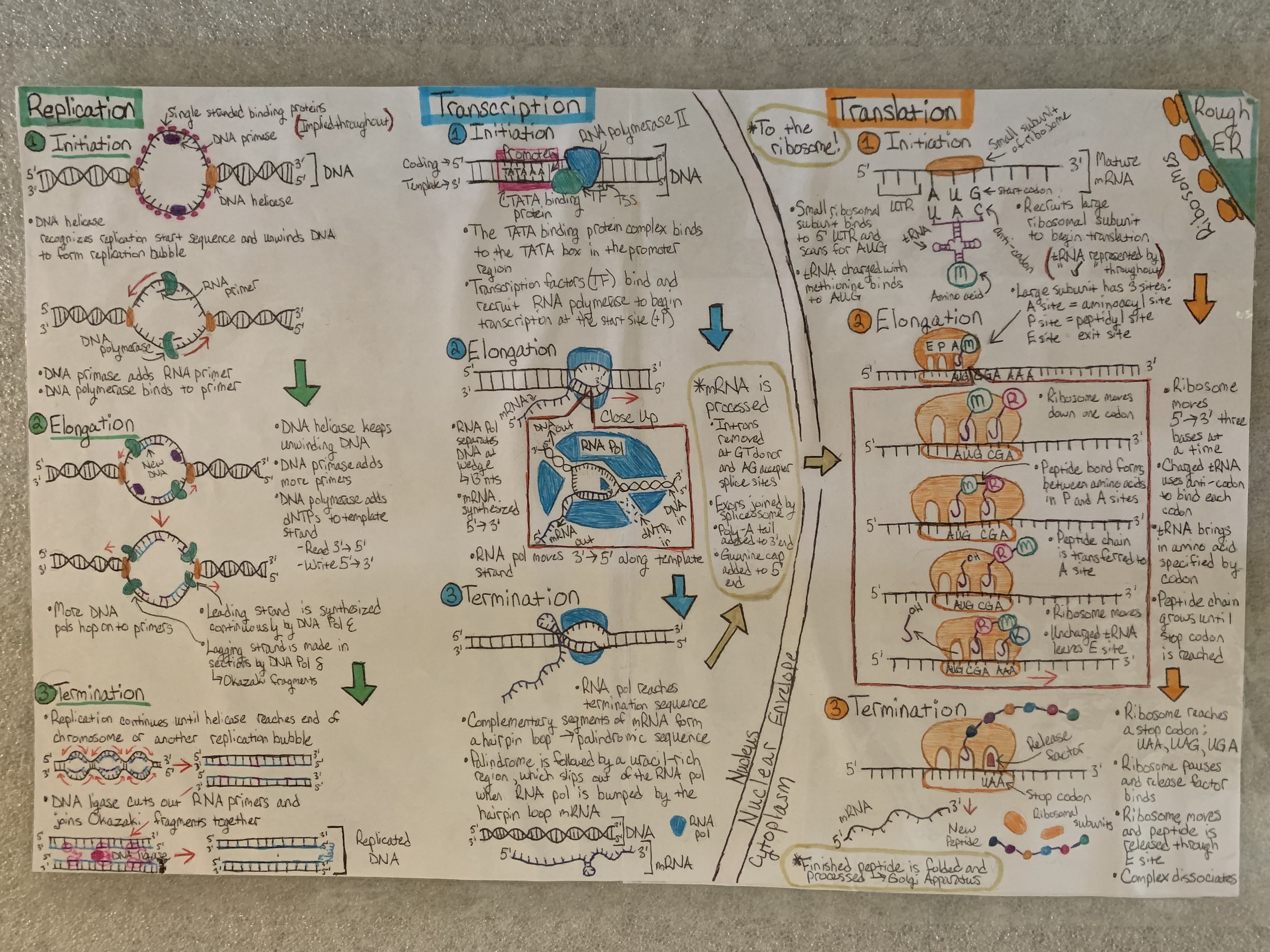
Primary image: Molecular View of the Central Dogma. This drawing was produced by a student at Bloomsburg University’s Human Genetics course for Part 1 of this lesson.

Rob Levenson onto Biotechnology
@
on