Creating a hands-on lab that conveys important information while simultaneously allowing for student autonomy can be difficult. This is particularly true for the field of microbiology, in which labs often rely on “recipe-style” instructions and materials that can be difficult to scale up for larger class sizes. For these reasons, microbiology concepts are often left out of introductory biology labs, the ramifications of which have been made apparent during the recent COVID-19 virus pandemic. Fundamental microbiology concepts, e.g., the prevention of communicable diseases, are important to teach in introductory biology classrooms – often a student's only exposure to biology in their academic careers – in order to create a healthier community as a whole. Therefore, this general biology lab introduces an active-learning microbiology lab that teaches students about the microbial world. Students are first introduced to the three major types of symbioses and apply these concepts to microbial organisms on a symbiotic continuum. Next, the students are given examples of mutualistic bacteria, i.e., the human microbiome, through a mini lecture prepared by the instructor. The students are then introduced to examples of parasitic/pathogenic microbes that can interfere with human health and cause relatable diseases (e.g., diarrhea, STDs, and athlete’s foot). Students then apply this information through a short matching game before learning common practices used to prevent the spread of these pathogens, including an active learning exercise and video on how to wash their hands like healthcare professionals. Finally, students are asked to generate their own questions about microbes before working through a handout that guides the students through using the scientific method to address their questions. This exercise thus provides students with the autonomy to ask their own questions about microbes, design their own experiments, prepare growth media their own way, and present their findings in a way that is both scalable for large class sizes and reduces the burden of lab prep common for microbiology labs.
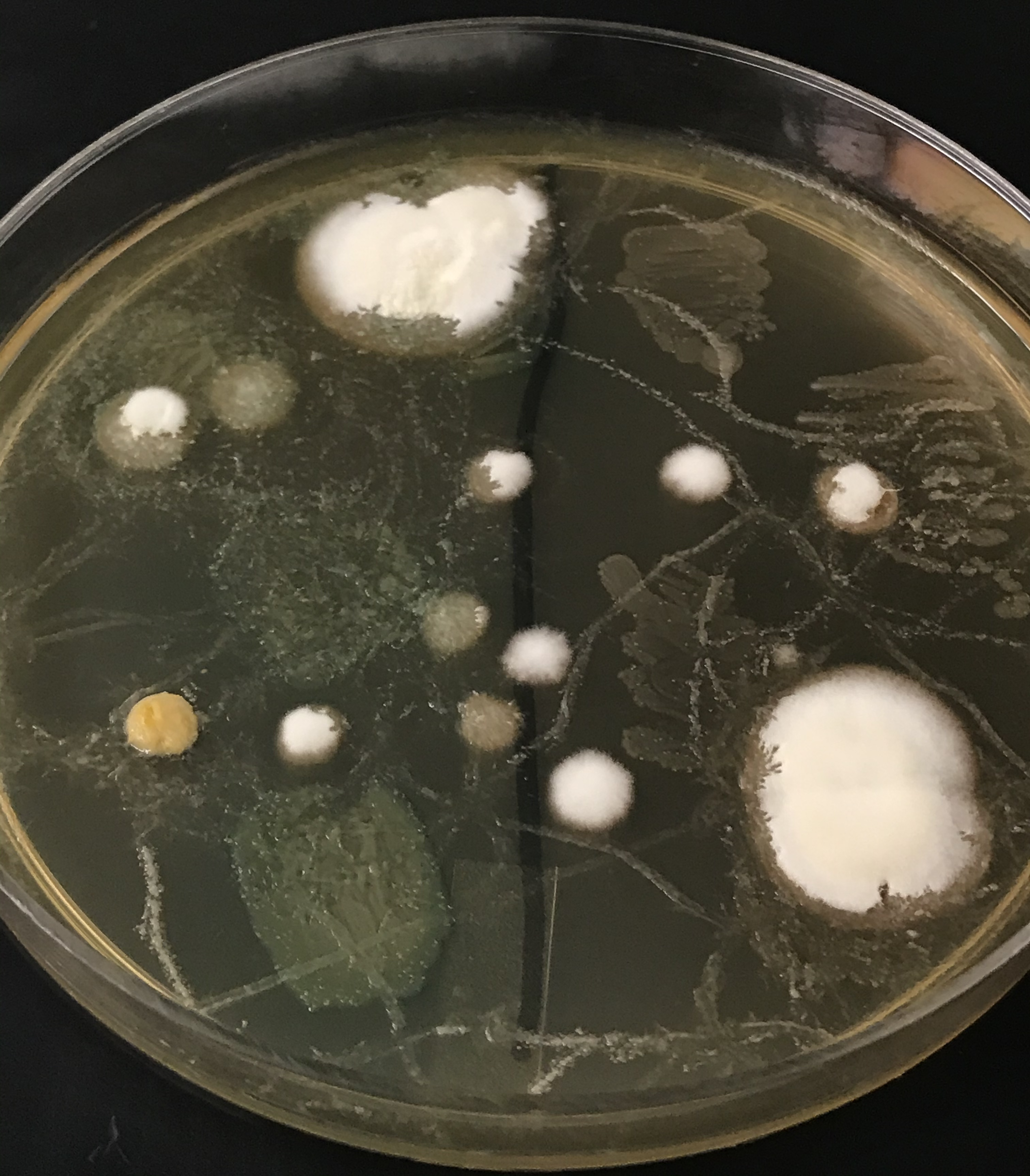
Primary image: Microbes sampled from the iPhone of a curious individual. Fungal colonies can be seen as fuzzy, white or colorful mounds while bacteria appear as opaque, smooth streaks on the media.