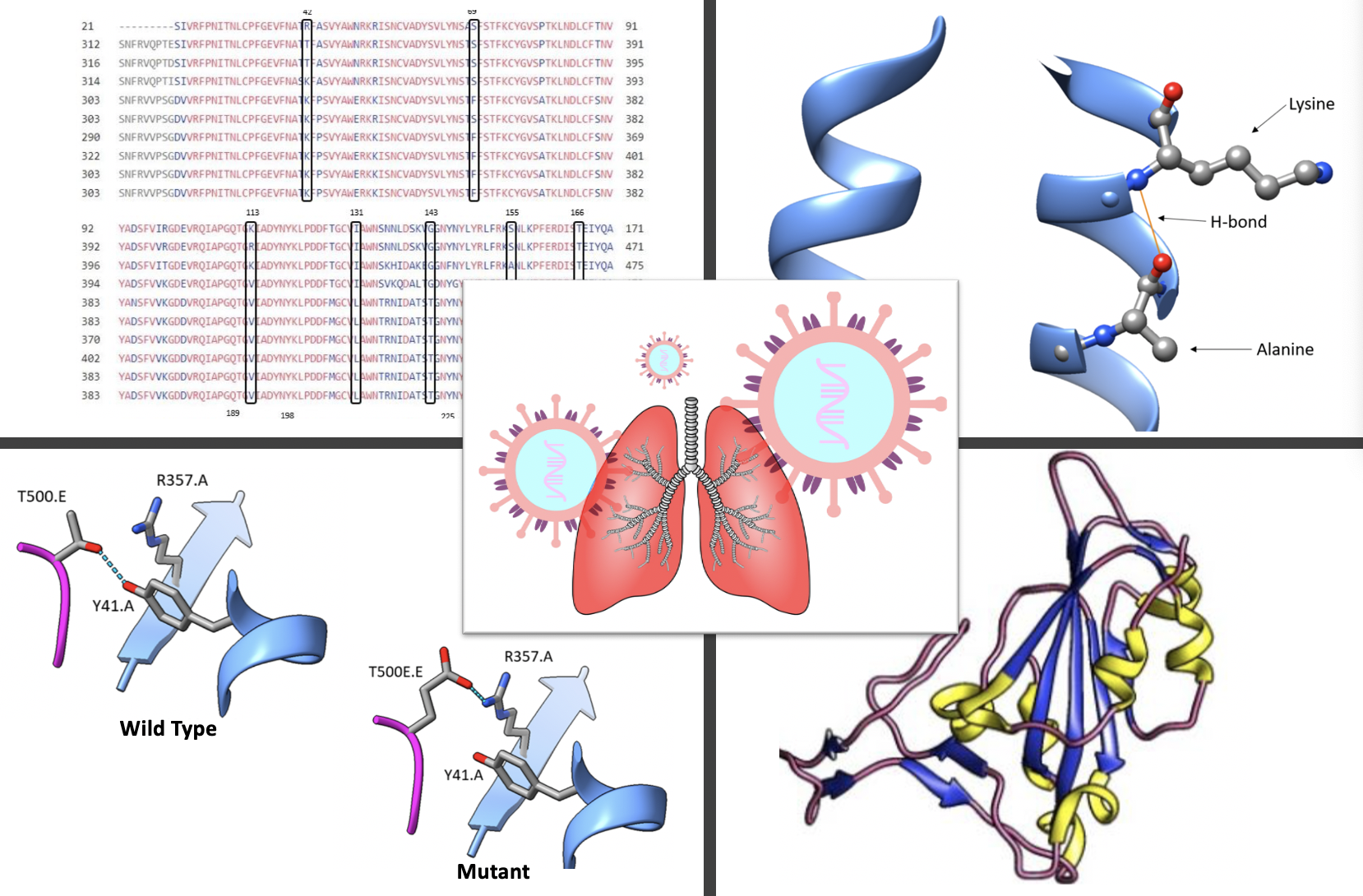
The relationship between protein structure and function is a foundational concept in undergraduate biochemistry. We find this theme is best presented with assignments that encourage exploration and analysis. Here, we share a series of four assignments that use open-source, online molecular visualization and bioinformatics tools to examine the interaction between the SARS-CoV-2 spike protein and the ACE2 receptor. The interaction between these two proteins initiates SARS-CoV-2 infection of human host cells and is the cause of COVID-19. In assignment I, students identify sequences with homology to the SARS-CoV-2 spike protein and use them to build a primary sequence alignment. Students make connections to a linked primary research article as an example of how scientists use molecular and phylogenetic analysis to explore the origins of a novel virus. Assignments II through IV teach students to use an online molecular visualization tool for analysis of secondary, tertiary, and quaternary structure. Emphasis is placed on identification of noncovalent interactions that stabilize the SARS-CoV-2 spike protein and mediate its interaction with ACE2. We assigned this project to upper-level undergraduate biochemistry students at a public university and liberal arts college. Students in our courses completed the project as individual homework assignments. However, we can easily envision implementation of this project during multiple in-class sessions or in a biochemistry laboratory using in-person or remote learning. We share this project as a resource for instructors who aim to teach protein structure and function using inquiry-based molecular visualization activities.
Primary image: Exploration of SARS-CoV-2 spike protein: student generated data from assignments I - IV. Includes examples of figures submitted by students, including a sequence alignment and representations of 3D protein structure generated using UCSF Chimera. The primary image includes student generated data and a cartoon from Pixabay, an online repository of copyright free art.